Endoglycoceramidase (EGCase, EC 3.2.1. 123) is capable of hydrolyzing the glycosidic linkage between the oligosaccharides and ceramide of various glycosphingolipids (GSLs) (Fig. 1). Three isoforms of EGCase differing in specificity have been found in Rhodococcus equi 1) and cloned 2) 3) 4). EGCase I and II hydrolyze all species of GSLs except for cerebrosides, sulfatide and 6-gala-series GSLs 1). Globo-series GSLs and fucosyl GM1a are hydrolyzed by EGCase I much faster than EGCase II 1). In contrast to EGCase I and II, EGCase III (endogalactosylceramidase, EGALC) hydrolyzes 6-gala-series GSLs and digalactosyl-diacylglycerol (DGDG) 3) 5). The specificity of EGCase is summarized in Table 1. Using EGCase, intact oligosaccharides and ceramide can be obtained from various GSLs; however one should consider which isoform of EGCase is suitable for an experiment. EGCase II is now available from TAKARA BIO INC.
EGCase is useful for analyzing the structure of various GSLs. For example, the released oligosaccharides can be labeled fluorescently by aromatic amine-based reagents 6)–10), and quantitatively determined by HPLC, TLC, and capillary electrophoresis (CE) with high sensitivity. Recently, a sensitive, rapid, and quantitative GSL-glycome analysis was developed, in which GSLs were hydrolyzed by EGCase, and the oligosaccharides released were subjected to glycoblotting and finally determined by MALDI-TOF MS 11).
Ceramide portions of 6-gala-series GSLs are exchanged with a fluorescent ceramide, various alkanols and non-ionic detergents such as Triton X-100 by a transglycosylation reaction of EGALC, generating fluorescent GSLs, alkyloligosaccahrides and Triton-oligosaccharides, respectively 12). |
| Category | Glycolipids and related compounds |
| Protocol Name | Degradation of Glycolipids by Endoglycoceramidase |
Authors
 |
Ishibashi, Yohei
*
Department of Bioscience and Biotechnology, Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University
Ito, Makoto
Department of Bioscience and Biotechnology, Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
Triton X-100 (Sigma-Aldrich, St. Louis, MO) |
| ● |
Pre-coated Silica gel 60 TLC plates (Merck Millipore, Billerica, MA) |
| ● |
|
| ● |
Sodium acetate buffer, pH 5.5 |
| ● |
Chloroform (Nacalai Tesque Inc., Kyoto, Japan) |
| ● |
Methanol (Nacalai Tesque, Inc.) |
| ● |
EGCase II (Recombinant enzyme expressed in E. coli is commercially available from Takara Bio Inc., Otsu, Japan) |
| ● |
|
| ● |
|
|
Instruments
 |
| ● |
Shimadzu CS-9300 TLC chromatoscanner (Shimadzu Corp., Kyoto, Japan) |
| ● |
Speed vac concentrator (Thermo Fisher Scientific Inc., Waltham, MA) |
| ● |
|
| ● |
|
|
| Methods |
|
1. |
Degradation of GSLs by EGCase |
| 1) |
Evaporate 2 nmol of each GSL in an Eppendorf tube with a speed bac concentrator when the sample is dissolved in an organic solvent. |
Comment 0
|

|
| 2) |
Dissolve the sample in 50 mM sodium acetate buffer, pH 5.5 containing 0.1% (w/v) Triton X-100, and sonicate for 10 sec. |
Comment 0
|

|
| 3) |
Add 0.5 mU of EGCase I or EGCase II for lacto- and ganglio-series GSLs, 10 mU of EGCase I for globo-series GSLs, and 1 mU of EGALC for 6-gala series GSLs. The volume of the reaction mixture should be 10–50 μL. One milliunit of enzyme is defined as that capable of catalyzing the hydrolysis of 1 nmol of GM1a per min. |
Comment 0
|

|
| 4) |
Incubate the reaction mixture at 37°C for 1 h for the hydrolysis of ganglio- and 6-gala series GSLs and for 10 h for the hydrolysis of lacto- and globo-series GSLs. Stop the reaction by heating in a boiling water bath for 5 min. |
Comment 0
|

|
| 6) |
Develop the TLC plate with chloroform/methanol/0.02% CaCl2 (5/4/1, v/v/v) for globo-, ganglio-, and lacto-series GSLs and chloroform/methanol/0.02% CaCl2 (2/3/1, v/v/v) for 6-gala-series GSLs. |
Comment 0
|
|
|
|
2. |
Detection of products by TLC chromatoscanner. |
| 1) |
Visualize the GSLs remaining and oligosaccharides generated by spraying the TLC plate with orcinol-H2SO4 reagent. |
Comment 0
|

|
| 2) |
Scan the band intensities of TLC using a Shimadzu CS-9300 chromatoscanner with the reflection mode set at 540 nm. |
Comment 0
|

|
| 3) |
Calculate the extent of hydrolysis as follows: hydrolysis (%) = (peak area for oligosaccharide) × 100 / (peak area for oligosaccharide + peak area for remaining substrate). |
Comment 0
|
|
|
| Notes | The specificity of EGCase I is somewhat different from that of EGCase II as shown in Table I. Kinetic study revealed that the Michaelis constant (Km) toward GM1a (ganglio series) and Gb3Cer (globo series) of EGCase I is almost the same of that of EGCase II (0.4 mM). On the other hand, rate of the turnover number (kcat) of EGCase I for GM1a and Gb3Cer is 22- and 1,209-fold that of EGCase II, respectively. The nonionic detergent Triton X-100 enhances the activity of EGCase I, II and EGALC at a concentration of 0.1%. No metal ions are required for the reactions of these enzymes. Not only 6-gala series GSLs but also DGDG can be hydrolyzed by EGALC 4).
[Reagents]
*If you need EGCase I and EGALC, please contact Dr. Makoto Ito at Kyushu University. |
| Figure & Legends |
Figure & Legends 

Fig. 1. Action points of EGCase I, II and III (EGALC) on GSLs.

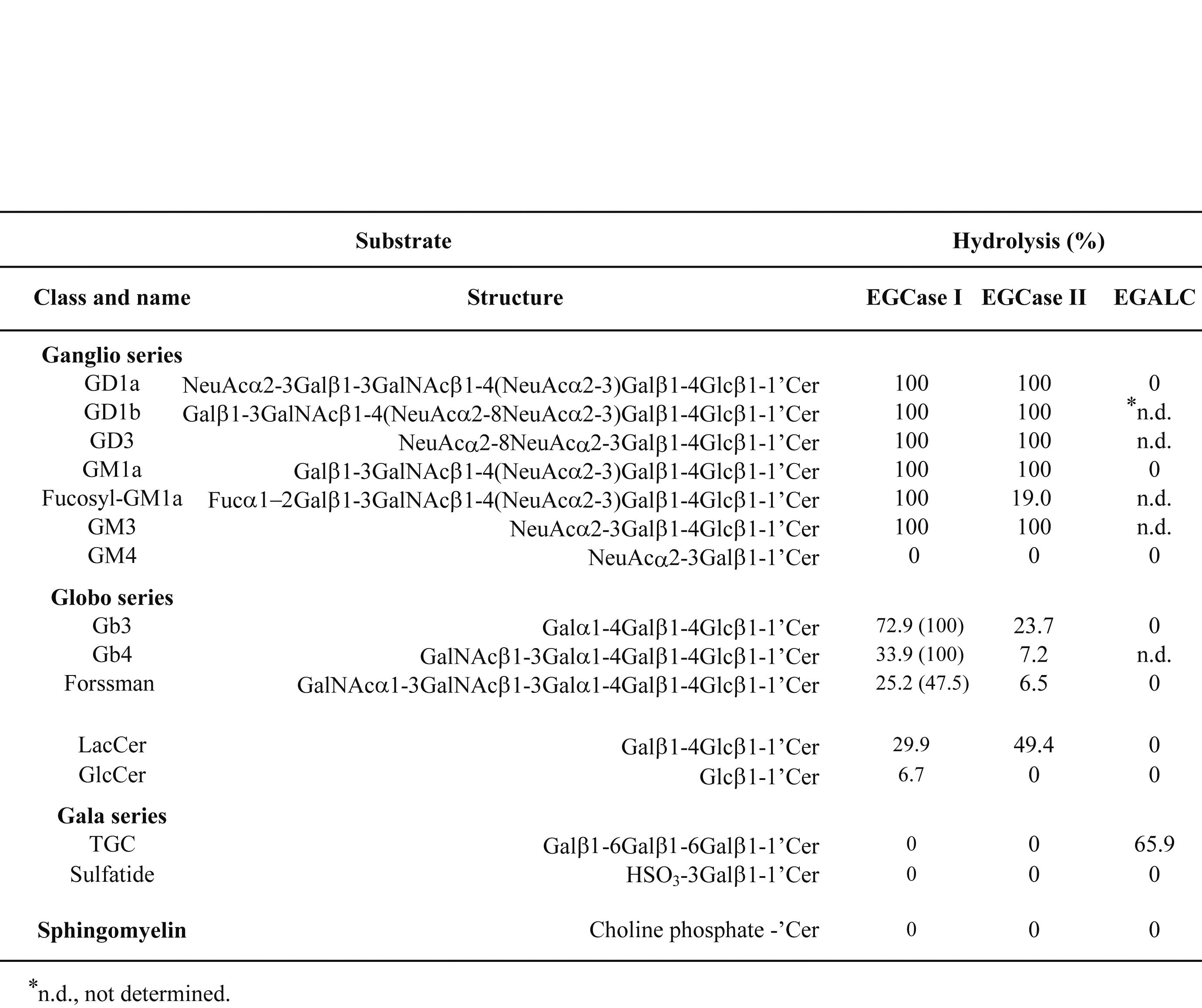
Table 1. Substrate specificities of EGCase I, II and III (EGALC).
Various GSLs (2 nmol) were incubated at 37°C for 12 h with 1 mU (10 mU) of enzyme in 20 mL of 50 mM sodium acetate buffer, pH 5.5, containing 0.1% Triton X-100. Values for EGALC were from ref. 3. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2016-01-19 14:41:37 |
- Ito, M., and Yamagata, T. (1989) Purification and characterization of glycosphingolipid-specific endoglycosidases (endoglycoceramidases) from a mutant strain of Rhodococcus sp. Evidence for three molecular species of endoglycoceramidase with different specificities. J Biol Chem. 264, 9510–9519 [PMID : 2722847]
- Izu, H., Izumi, Y., Kurome, Y., Sano, M., Kondo, A., Kato, I., and Ito, M. (1997) Molecular cloning, expression, and sequence analysis of the endoglycoceramidase II gene from Rhodococcus species strain M-777. J Biol Chem. 272, 19846–19850 [PMID : 9242646]
- Ishibashi, Y., Nakasone, T., Kiyohara, M., Horibata, Y., Sakaguchi, K., Hijikata, A., Ichinose, S., Omori, A., Yasui, Y., Imamura, A., Ishida, H., Kiso, M., Okino, N., and Ito, M. (2007) A novel endoglycoceramidase hydrolyzes oligogalactosylceramides to produce galactooligosaccharides and ceramides. J Biol Chem. 282, 11386–11396 [PMID : 17244618]
- Ishibashi, Y., Kobayashi, U., Hijikata, A., Sakaguchi, K., Goda, H., Tamura, T., Okino, N., and Ito, M. (2012) Preparation and characterization of endoglycoceramdiase I, applicable to the comprehensive analysis of glycosphingolipids, using a rhodococcal expression system. J. Lipid Res. 53, 2242–2251 [PMID : 22798689]
- Ishibashi, Y., Nagamatsu, Y., Meyer, S., Imamura, A., Ishida, H., Kiso, M., Okino, N., Geyer, R., and Ito, M. (2009) Transglycosylation-based fluorescent labeling of 6-gala series glycolipids by endogalactosylceramidase. Glycobiology 19, 797–807 [PMID : 19389917]
- Higashi, H., Ito, M., Fukaya, N., Yamagata, S., and Yamagata, T. (1990) Two-dimensional mapping by the high-performance liquid chromatography of oligosaccharides released from glycosphingolipids by endoglycoceramidase. Anal Biochem. 186, 355–362 [PMID : 2363510]
- Ohara, K., Sano, M., Kondo, A., and Kato, I. (1991) Two-dimensional mapping by high-performance liquid chromatography of pyridylamino oligosaccharides from various glycosphingolipids. J Chromatogr. 586, 35–41 [PMID : 1806553]
- Basu, S. S., Dastgheibhosseini, S., Hoover, G., Li, Z. X., and Basu, S. (1994) Analysis of glycosphingolipids by fluorophore-assisted carbohydrate electrophoresis using ceramide glycanase from Mercenaria mercenaria. Anal Biochem. 222, 270–274 [PMID : 7856860]
- Wing, D. R., Garner, B., Hunnam, V., Reinkensmeier, G., Andersson, U., Harvey, D. J., Dwek, R. A., Platt, F. M., and Butters, T. D. (2001) High-performance liquid chromatography analysis of ganglioside carbohydrates at the picomole level after ceramide glycanase digestion and fluorescent labeling with 2-aminobenzamide. Anal Biochem. 298, 207–217 [PMID : 11700975]
- Neville, D. C., Coquard, V., Priestman, D. A., te Vruchte, D. J., Sillence, D. J., Dwek, R. A., Platt, F. M., and Butters, T. D. (2004) Analysis of fluorescently labeled glycosphingolipid-derived oligosaccharides following ceramide glycanase digestion and anthranilic acid labeling. Anal Biochem. 331, 275–282 [PMID : 15265733]
- Fujitani, N., Takegawa, Y., Ishibashi, Y., Araki, K., Furukawa, J., Mitsutake, S., Igarashi, Y., Ito, M., and Shinohara, Y. (2011) Qualitative and quantitative cellular glycomics of glycosphingolipids based on rhodococcal endoglycosylceramidase-assisted glycan cleavage, glycoblotting-assisted sample preparation, and matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry analysis. J Biol Chem. 286, 41669–41679 [PMID : 21965662]
- Ishibashi, Y., Kiyohara, M., Okino, N., and Ito, M. (2007) Synthesis of fluorescent glycosphingolipids and neoglycoconjugates which contain 6-gala oligosaccharides using the transglycosylation reaction of a novel endoglycoceramidase (EGALC). J Biochem. 142, 239–246 [PMID : 17567653]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Ishibashi, Yohei,
Ito, Makoto,
(2016). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.7,4,2025 .
How to Cite this Work in Website:
Ishibashi, Yohei,
Ito, Makoto,
(2016).
Degradation of Glycolipids by Endoglycoceramidase.
Retrieved 7,4,2025 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t175.
html source
Ishibashi, Yohei,
Ito, Makoto,
(2016).
<b>Degradation of Glycolipids by Endoglycoceramidase</b>.
Retrieved 4 7,2025 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t175" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t175</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|