Format Type: SMILES, MOL, SDF, CML, SYBYL
Format Type: SMILES, MOL, SDF, CML, SYBYL
Cautions (Close)
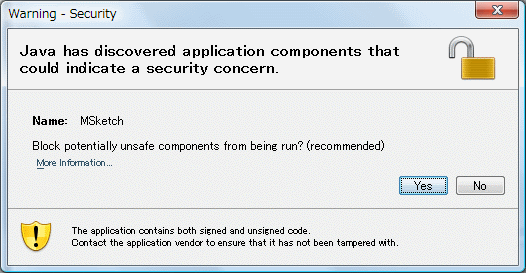
At this site, when a re-display and change of a screen are carried out, the following screens may be displayed.
Please choose "No" at this time.
When "Yes" is chosen, there is a case where it becomes impossible to use a part of functions.
|
Objective
In the Japan Consortium for Glycobiology and Glycotechnology DataBase (JCGGDB) which is open to the public,
there are databases that integrate structural information of glyco-related compounds,
including JMSDB which stores structural information of such basic monosaccharides as glucose and fucose,
JIGSDB which stores more complex glycan structural information,
and a database which stores structural information of glycosides including inhibitors such as glycosyltransferase and glycosidase.
However, because glyco-related compounds stored in JCGGDB are composed of not just monosaccharides but also aglycons (e.g. steroid, phenol, flavonoid),
there have been only limited uses for glycobiologists who are accustomed to searching for structures with CFG monosaccharide symbols.
To solve this problem, we have developed GlycoChemExplore,
by which molecular structures of compounds can be searched for with chemical structural formula of aglycons as well as glycans.
(Note: GlycoChemExplore does not cover glycans in JIGSDB of which bonding positions and repetition of structures are uncertain.)
Brief overview
GlycoChemExplore is a search system in which molecular structures of monossacharides,
glycans and a variety of other substances that have in part glycan structures,
are made into queries and searched for.
This system enables the followings
- It can search for glycans by molecular structures and glyco-related compounds by organic chemistry.
- It can search for glycan structures of which the conformation is defined, such as glycoside bonding and anomeric.
- Besides the JCGGDB data, it stores data of organic compounds in the GlycoNAVI Database which has been accumulating data with the help of a working group in which Noguchi Institute played a central role.
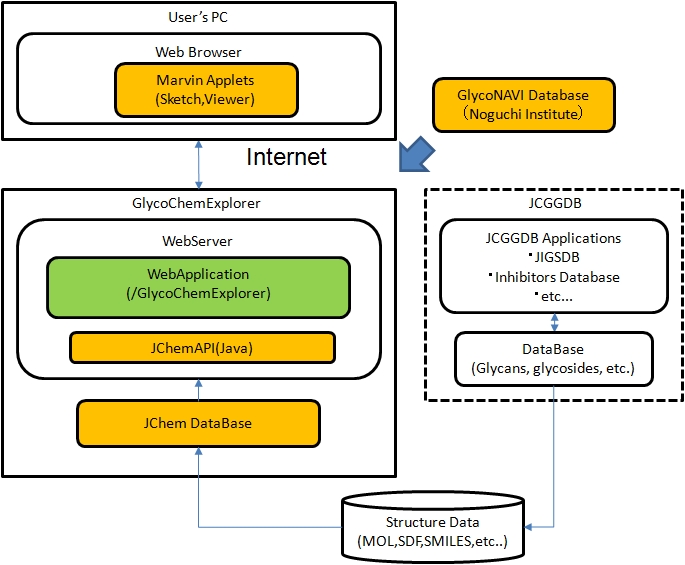
Fig. System Structure
As of March 2013, the data extracted from the following databases are registered as search targets.
Also, data are updated on a timely basis after each of the databases is updated.
Acknowledgement
GlycoChemExplorer was developed with the support of the Integrated Database Project by the Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Updating of data and improvement of functions are made with the help of the Integrated Database Program sponsored by Japan Science and Technology Agency (JST) and National Bioscience Database Center (NBDC).
MEXT integrated Database Project, web site : http://lifesciencedb.mext.go.jp/
NBDC integrated Program, web site : http://biosciencedbc.jp/en/
Disclaimer
The Research Center for Medical Glycoscience of the National Institute of Advanced Industrial Science and Technology (AIST) makes all the efforts to provide correct and up-to-date information,
but it accepts no liability for the accuracy of information, or the correctness, completeness or usefulness of data available in this website.
How to use the system
There are four ways to use this system:
・Searching by drawing a structure (Draw a structure)
・Displaying a structure by specifying an identifier, and then editing (Draw a structure by Identifier)
・Describing a structure by text format of chemical structures file such as SMILES, MOL, SDF, CML and SYBYL, and displaying it to edit (Draw a structure by “Copy & paste”)
・Describing a structure by text format of chemical structures file, then converting it to another format (Convert)
Note: The selected function is shown by a solid line (In the example below, “Samples” is selected).

Note: MarvinSketch (ChemAxion Ltd.) is used for drawing structures, so please refer to MarvinSketch GUI for more details.
The following five search conditions can be set:
| Search condition |
Meaning |
| PERFECT |
Search for structures that match perfectly, including conformation. |
| EXACT |
Search for structures that match exactly but allow for vagueness in conformation. |
| SUBSTRUCTURE |
Search for structures that match partly. |
| SIMILARITY |
Search for structures that are similar. |
The following formats can be used for reading and converting:
| Format |
Related URL |
| Molfile,SDfile |
http://accelrys.com/products/informatics/cheminformatics/ctfile-formats/no-fee.php |
| CML |
http://cml.sourceforge.net/ |
| SMILES |
http://www.daylight.com/dayhtml/doc/theory/theory.smiles.html |
| SYBYL mol |
https://www.chemaxon.com/marvin/help/formats/sybyl-doc.html |
| PDB format |
https://www.chemaxon.com/marvin/help/formats/pdb-doc.html |
| XYZ format |
https://www.chemaxon.com/marvin/help/formats/xyz-doc.html |
How to operate on each display screen is described below:
(1)Draw a structure
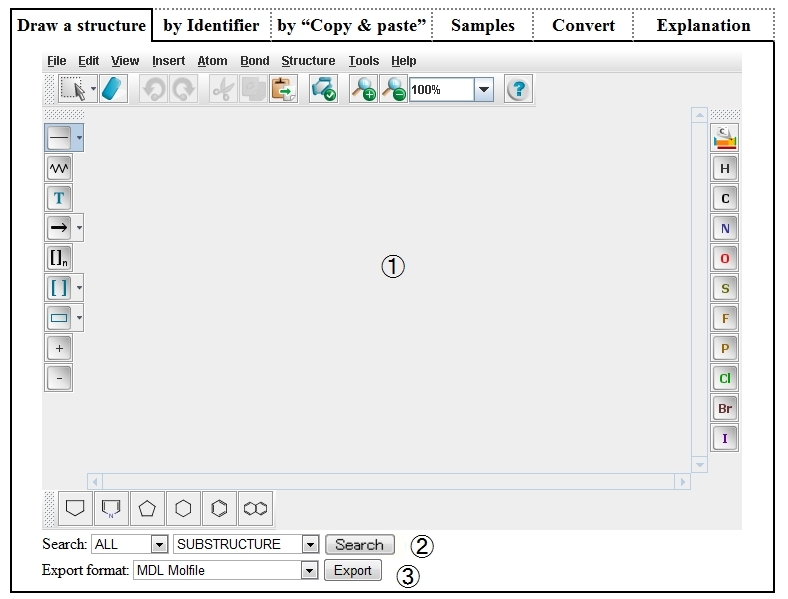
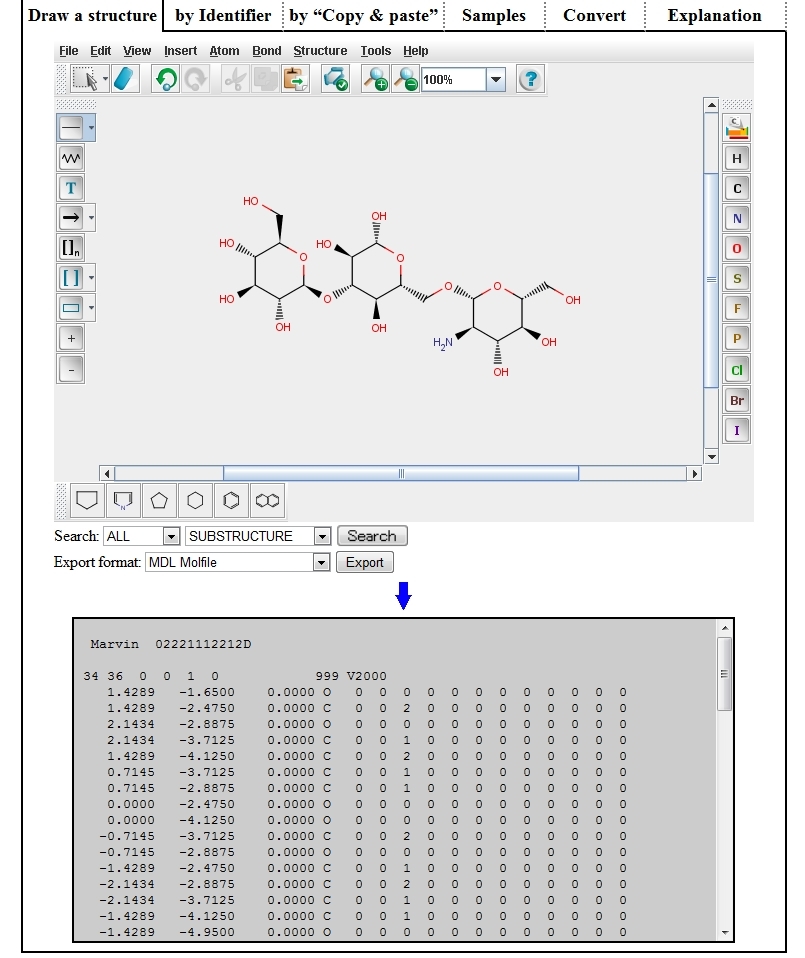
| ➀ | On Structure Editor, draw a structure by operating the mouse and buttons, or by reading a file in which MOL, SMILES, etc. are described. |
|
| ② | Select a data format and search condition, and click the “Search” button, and the search result will be displayed on another tab (or window) of the browser . |
|
| ③ | Select an output format and click the “Export” button, and the following textbox will appear and the text described in the specified format will be displayed. |
(2)(Draw a structure)by Identifier
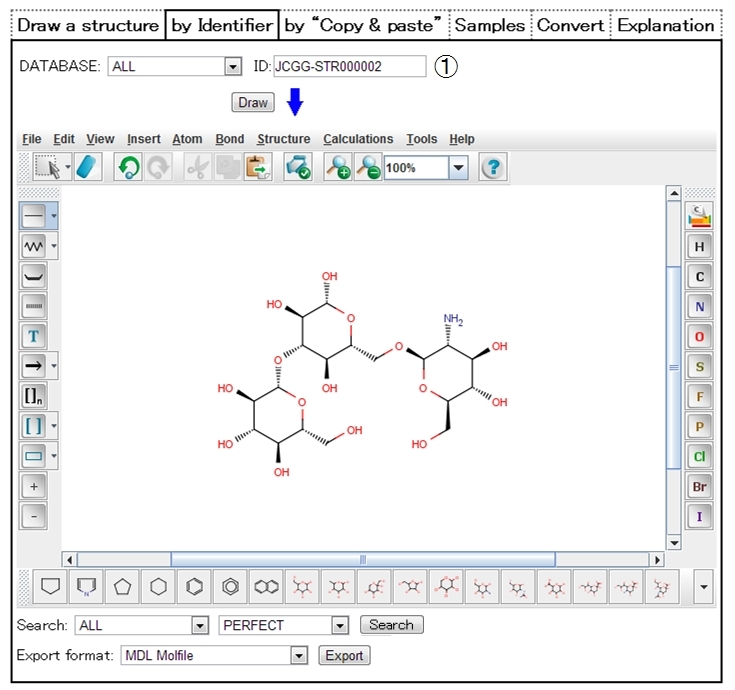
| ➀ | Enter a JCGGDB ID (e.g. JCGG-STR000002,JMS000) on the ID column, select a data format if necessary, and click the “Draw” button, and the structure of the matched data will be displayed on Structure Editor. |
(3)(Draw a structure)by “Copy & paste”
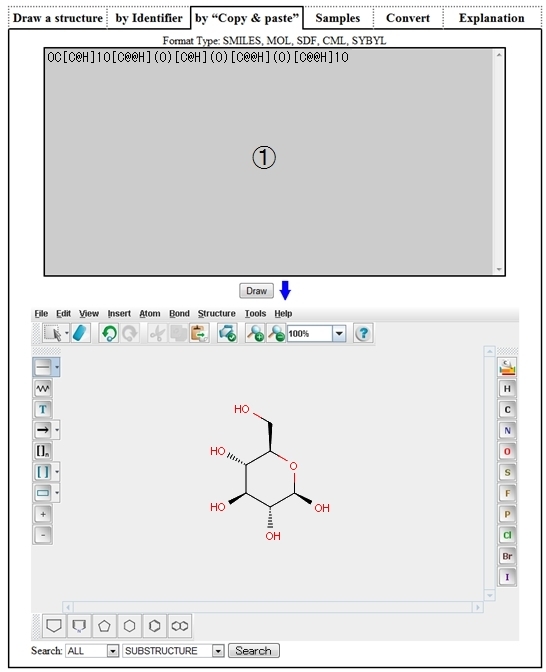
| ➀ | Describe a structure by SMILE, MOL, etc. on the above textbox and click the “Draw” button, and the structure of the matched data will be displayed on Structure Editor. |
(4)Convert
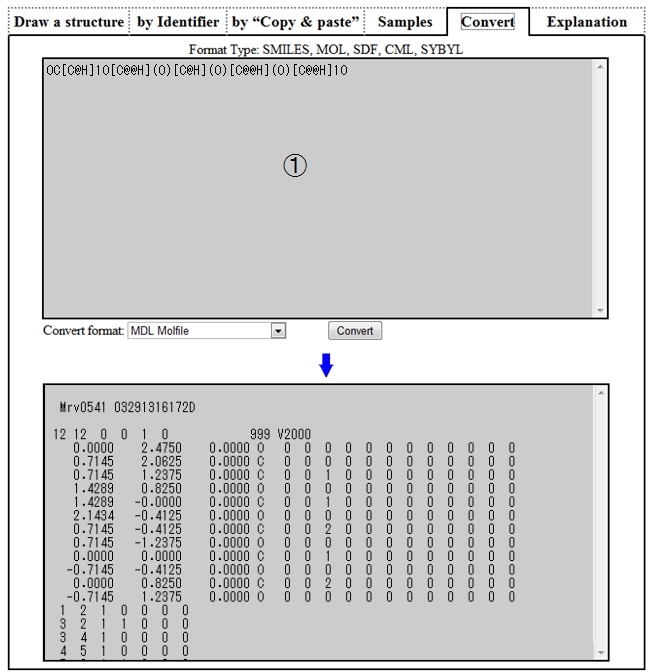
| ➀ | Describe a structure by SMILES, MOL, etc. on the above textbox, select a convert format and click the “Convert” button, and the text described in the specified format will be displayed on the textbox below. |
If you have any questions, please contact JCGGDB Help Desk at Research Center for Medical Glycoscience(RCMG), National Institute of Advanced Industrial Science and Technology (AIST).
Mail address: 
|
|
|
|
|
Search:
|
|
Export format:
|
|
|
|


