Accumulating evidence implies important biological functions of glycosaminoglycans (GAGs). To clarify the structural basis of these biological activities, it is necessary to purify GAGs from target tissues and cells. Here we describe protocols for the preparation of GAGs from tissues on a relatively large scale and from cells or small amounts of tissue for microdetermination by HPLC. |
| Category | Glycosaminoglycans |
| Protocol Name | Preparation of glycosaminoglycans |
Authors
 |
Yamada, Shuhei
*
Department of Pathobiochemistry, Faculty of Pharmacy, Meijo University
Toyoda, Hidenao
Laboratory of Bio-analytical Chemistry, College of Pharmaceutical Sciences, Ritsumeikan University
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
Actinase E (Kaken Pharmaceutical Co., Ltd., Tokyo, Japan) |
|
Instruments
 |
| ● |
Speed Vac Concentrator (Thermo Fisher Scientific Inc., Waltham, MA) |
| ● |
Accell QMA Plus cartridge (Waters Corp., Milford, MA) |
| ● |
PD-10 column (GE Healthcare, Little Chalfont, UK) |
| ● |
Microcon® YM-10 (Merck Millipore, Billerica, MA) |
| ● |
Ultrafree MC (Durapore, 0.45 μm) (Merck Millipore) |
| ● |
Ultrafree MC DEAE (Merck Millipore) |
|
| Methods |
|
1. |
Preparation of GAGs from tissue |
| 1) |
Homogenize a tissue in ice-cold acetone. |
Comment 0
|

|
| 3) |
Suspend the dried material (25 g) in distilled water (40 mL). |
Comment 0
|

|
| 4) |
Inactivate contaminating glycosidases by boiling at 100˚C for 10 min. |
Comment 0
|

|
| 5) |
Digest the sample with actinase E (0.25 g) in a total volume of 65 mL of 0.1 M borate buffer, pH 8.0, containing 10 mM CaCl2 at 65˚C for 2 days. |
Comment 1
|

|
| 6) |
Treat the digest with 5% trichloroacetic acid at 4˚C for 1 h to precipitate residual proteins and peptides. |
Comment 0
|

|
| 7) |
Centrifuge at 3,500 rpm for 10 min to remove the precipitate. |
Comment 0
|

|
| 8) |
Remove trichloroacetic acid by extraction with an equivalent volume of diethylether 5 times. |
Comment 0
|

|
| 9) |
Neutralize the aqueous phase with 1 M Na2CO3. |
Comment 0
|

|
| 10) |
Precipitate the crude GAG with 80% ethanol containing 1% sodium acetate at 4˚C overnight. |
Comment 0
|

|
| 11) |
Centrifuge at 3,000 rpm for 10 min. |
Comment 0
|

|
| 12) |
Reconstitute the precipitate in distilled water. |
Comment 0
|

|
| 13) |
Load the sample onto a prepacked disposable PD-10 column equilibrated with H2O to be desalted. |
Comment 0
|

|
| 14) |
Lyophilize the flow-through fraction by Speed Vac. |
Comment 0
|

|
| 15) |
Quantify the yield of GAGs by the carbazole reaction. |
Comment 1
|

|
| 16) |
Load the crude GAG fraction (2.5 mg as GAG) on an Accell QMA Plus cartridge pre-equilibrated with 0.05 M phosphate buffer, pH 6.0, containing 0.15 M NaCl. |
Comment 1
|

|
| 17) |
Wash the column with 2 mL of the equilibration buffer. |
Comment 0
|

|
| 18) |
Elute GAGs from the column with 3 mL each of 0.05 M phosphate buffer, pH 6.0, containing 0.5 and 2.0 M NaCl. |
Comment 0
|

|
| 19) |
Dialyze the eluates against water using Microcon® YM-10 and concentrate them by Speed Vac. |
Comment 0
|
|
|
|
2. |
Preparation of GAGs for microdetermination |
| 1) |
Lyophilize samples (cells/small amount of tissues) over 16 h. |
Comment 0
|

|
| 2) |
Homogenize the samples (up to 107 cells/up to 20 mg of lyophilized tissue) in 1.0 mL of acetone cooled on ice. Stand for 15 min on ice, then centrifuge at 2,300 × g for 15 min at 4℃. Wash the precipitate with 1.0 mL of ice-cold acetone and dry under vacuum. |
Comment 0
|

|
| 3) |
Extract samples in 1.0 mL of GAG extraction solution (0.5% SDS, 0.1 M NaOH, and 0.8% NaBH4) for 16 h at room temperature with constant stirring. |
Comment 0
|

|
| 4) |
Neutralize samples with 200 μL of 1.0 M sodium acetate and 300 μL of 1.0 M HCl and centrifuge at 2,300 × g for 10 min at 4℃. Remove particulate matter from the supernatant with a 300-μm pore disposable filter column. |
Comment 0
|

|
| 5) |
Add 200 μL of 1.0 M HCl to the filtrate and remove insoluble materials by centrifugation at 2,300 × g for 10 min at 4˚C. Add 7 mL of ethanol saturated with sodium acetate to the supernatant and allow GAGs to precipitate for 16 h at 4˚C. |
Comment 0
|

|
| 6) |
Centrifuge at 2,300 × g for 15 min at 4℃ and remove the supernatant. Wash the precipitate once with 1 mL of 80% ethanol and once with 1 mL of ethanol. Dry the precipitate under vacuum. At this stage, the crude GAG pellets can be stored at −20℃ until needed. |
Comment 0
|

|
| 7) |
Dissolve the crude GAG pellets in 250 μL of water. |
Comment 0
|

|
| 8) |
Add 50 μL of 0.3 M sodium phosphate buffer, pH 6.0, to the crude GAG solution. |
Comment 0
|

|
| 9) |
Filter the mixture through an Ultrafree MC (Durapore, 0.45 μm). |
Comment 0
|

|
| 10) |
Add the filtrate to the Ultrafree MC DEAE insert pre-equilibrated with loading buffer (50 mM sodium phosphate buffer, pH 6.0) and spin at 5,000 × g for 1 min. Pass the sample over the membrane twice. |
Comment 0
|

|
| 11) |
Add 400 μL of loading buffer to the insert and spin at 5,000 × g for 1 min. |
Comment 0
|

|
| 12) |
Transfer the insert to a new microcentrifuge tube. Add 100 μL of elution buffer (50 mM sodium phosphate buffer, pH 6.0, containing 1.0 M NaCl) to the insert and spin at 5,000 × g for 1 min. Repeat 3 times. This fraction is the eluate. |
Comment 0
|

|
| 13) |
Add the eluate to Ultrafree MC (Biomax 5) and spin until the retentate is about 30 μL. |
Comment 0
|

|
| 14) |
Add 50 μL of distilled water to the retentate and spin. Repeat four times. |
Comment 0
|

|
| 15) |
Remove the retentate to a new micro tube and rinse the membrane four times with 20 μL of distilled water. Add the washes to the retentate. |
Comment 0
|

|
|
|
| Figure & Legends |
Figure & Legends 
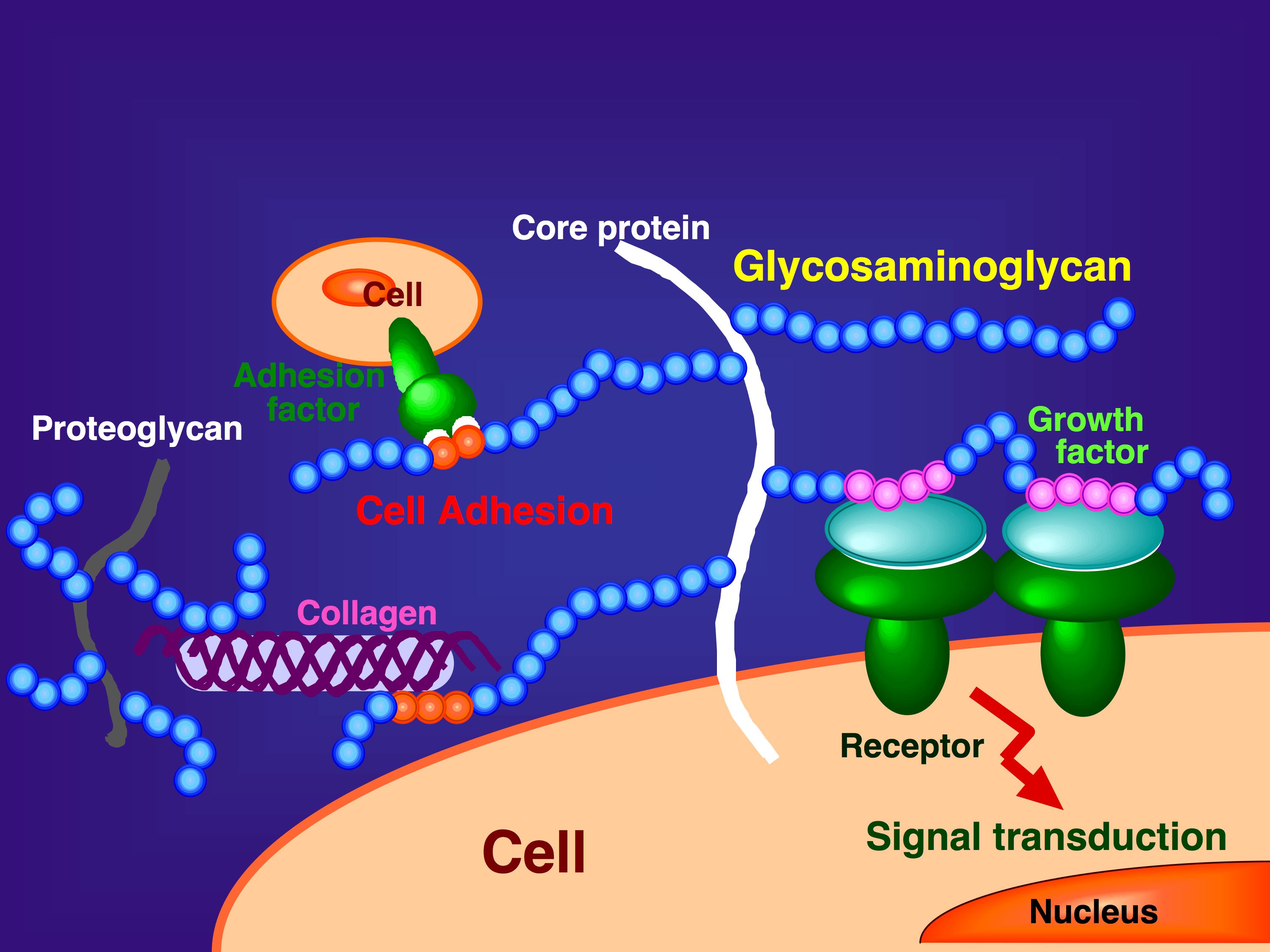
Fig. 1. Functions of GAGs on the cell surface and in the extracellular matrix
GAGs are distributed as side chains of proteoglycans at the cell surface or in the extracellular matrix of animal tissues. They play important roles in biological processes such as growth factor signaling, cell adhesion, and interactions with extracellular matrix components.


Fig. 2. Structure of GAGs
The structure of the repeating disaccharide region of hyaluronic acid (HA), chondroitin sulfate (CS), dermatan sulfate (DS), heparan sulfate (HS)/heparin (Hep), and keratan sulfate (KS) is shown. Possible sulfation positions are given in the structure of CS, DS, HS/Hep, and KS. An asterisk in the structure of HS/Hep indicates that the C5 position of the uronic acid residue may be epimerized. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2016-11-17 10:56:56 |
- Yamada, S., Onishi, M., Fujinawa, R., Tadokoro, Y., Okabayashi, K., Asashima, M., and Sugahara, K. (2009) Structural and functional changes of sulfated glycosaminoglycans in Xenopus laevis during embryogenesis. Glycobiology 19, 488–498 [PMID : 19190026]
- Toyoda, H., Kinoshita-Toyoda, A., and Selleck, S.B. (2000) Structural analysis of glycosaminoglycans in Drosophila and Caenorhabditis elegans and demonstration that tout-velu, a Drosophila gene related to EXT tumor suppressors, affects heparan sulfate in vivo. J Biol Chem 275, 2269–2275 [PMID : 10644672]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Yamada, Shuhei,
Toyoda, Hidenao,
(2016). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.3,5,2024 .
How to Cite this Work in Website:
Yamada, Shuhei,
Toyoda, Hidenao,
(2016).
Preparation of glycosaminoglycans.
Retrieved 3,5,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t15.
html source
Yamada, Shuhei,
Toyoda, Hidenao,
(2016).
<b>Preparation of glycosaminoglycans</b>.
Retrieved 5 3,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t15" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t15</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|