GMDB使用方法
GMDB内の様々な糖鎖のMSnのスペクトルと検体のスペクトルとを比較することにより、糖鎖構造を推定することができます。GMDBのスペクトル検索には、糖鎖組成から検索する方法と、前駆イオンのm/z値から検索する方法があります。
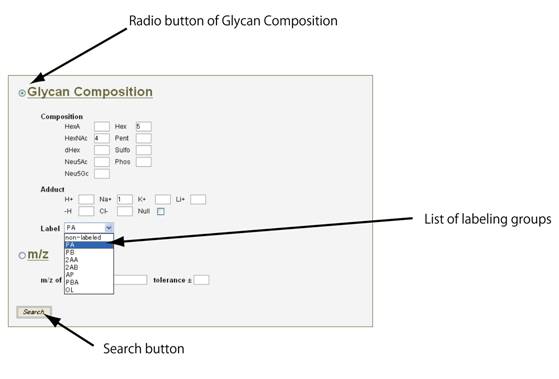
糖鎖組成から検索する場合は、”Glycan Composition”のラジオボタンをクリックしてください。
PA: pyridyl amino PB: pyrene butylic hydrazide 2AA: 2-amido-benzoic acid 2AB: 2-amido-benzamide AP: aminopyrene PBA: pyrene butylamine OL: alditol
最後に、”Search” ボタンをクリックしてください。
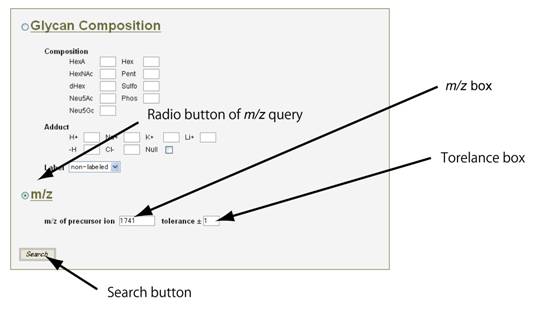
m/z値から検索する場合は、”m/z” のラジオボタンをクリックします。 そして、比較したいスペクトルの前駆イオンの m/z 値を入力します。次に、”tolerance”の欄に適当な数字を入力してください。通常、0.5から1.0までの数字を入れることによって、関連スペクトルを見つけることができます。
最後に、”Search”ボタンをクリックしてください。
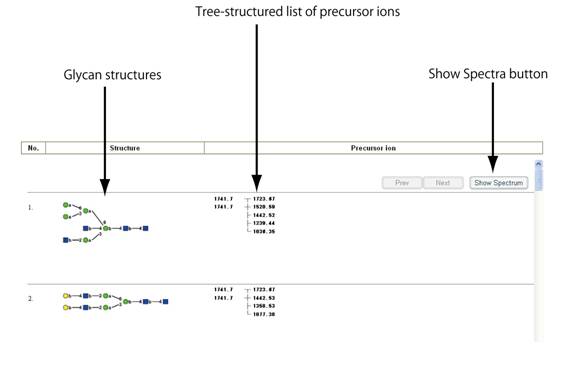
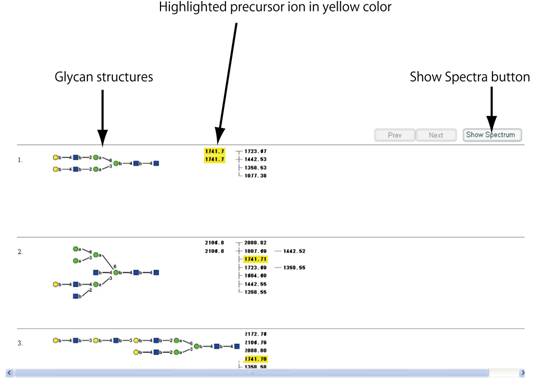
Searchボタンを押すと、「Spectral Database Search Result」のウィンドウが開き、検索した条件に合致するスペクトルを与えた糖鎖構造が表示されます。表示されている糖鎖に関してGMDB内に格納されているMSnスペクトルのリストは、前駆イオンのm/z値のツリー構造によって表示されます。m/z値で検索した場合、合致する前駆イオンは黄色で強調されています。参照したいスペクトルの前駆イオンのm/z値をクリックして選択してください。選択をキャンセルしたい場合は、黄色で強調された数字を再度クリックすると元に戻ります。選択したスペクトルを表示するには、「Show Spectra」のボタンをクリックしてください。
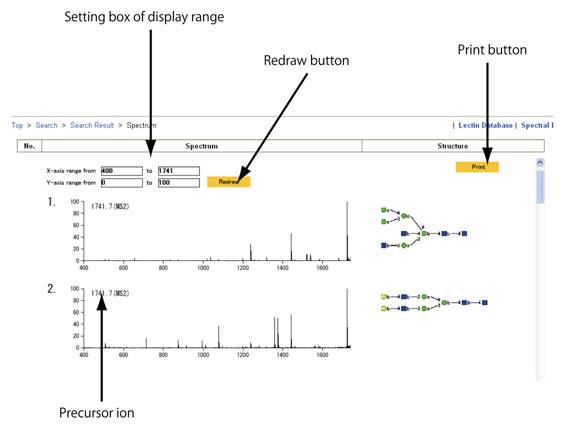
スペクトルは、そのスペクトルを与えた糖鎖構造と共にウィンドウに現れます。前駆イオンの値もスペクトルの左上に示されます。MS4スペクトルの場合は、断片化経路も示されます。左上に表示されているディスプレイ範囲設定ボックスに適切な値を入力した後に、「Redraw」ボタンをクリックすることにより、拡大されたスペクトルを表示することもできます。表示されたスペクトルを印刷したい場合は、「Print」のボタンをクリックしてください。
GGDBのブラウジング情報 ウェブブラウザ(インターネット・エクスプローラー、FireFox、およびSafariなど)でGMDBの情報にアクセスして、見ることができます。 |