EDEMs (ER degradation enhancing α-mannosidase-like protein) constitute a subgroup of Class I α1,2-mannosidases (glycosylhydrolase family 47). EDEMs are involved in ERQC (ER quality control) or ERAD (ER-associated degradation) of glycoproteins. Saccharomyces cerevisiae has one homologue protein named Htm1p/Mnl1p (Jakob et al., 2001), and mammals have three homologues of EDEM1, 2, and 3 (Hirao et al., 2006; Hosokawa et al., 2001) (Fig. 1). Htm1p and EDEM1 were postulated as Man8 lectins when they were cloned, because enzyme activities were not detected. However, recent works have suggested that Htm1p (Clerc et al., 2009), EDEM1 (Hosokawa et al., 2010) and EDEM3 (Hirao et al., 2006) have processing α-mannosidase activity in vivo within the cells. Here, protocols to isolate tagged EDEMs expressed in mammalian cells and to detect their binding with ERAD substrates in the cells are described. |
| Category | Sugar binding proteins |
| Protocol Name | Isolation and binding assay of EDEMs |
Authors
 |
Hosokawa, Nobuko
Department of Molecular and Cellular Biology, Institute for Frontier Medical Sciences, Kyoto University
|
| KeyWords |
|
Reagents
 |
| ● |
DMEM without methionine (& without cysteine) (e.g. Invitrogen/Life Technologies, Carlsbad, CA) |
| ● |
Dialyzed FBS (fetal bovine serum) |
| ● |
PBS (Dulbecco’s phosphate buffered saline) |
| ● |
[35S]Methionine or a mixture of [35S]Methionine & [35S]Cysteine ([35S] Protein Labeling Mixture) (e.g. PerkinElmer, Waltham, MA) |
| ● |
Protein A Sepharose or Protein G Sepharose (e.g. GE Healthcare, Little Chalfont, UK) |
| ● |
1% NonidetP-40 (NP-40) lysis buffer; 1% NP-40, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl |
| ● |
High ionic extraction buffer; 1% NP-40, 50 mM Tris-HCl, pH 7.5, 400 mM NaCl |
| ● |
Protease inhibitors (e.g. 2 mM NEM, 0.2 mM AEBSF, 1μg/mL Leupeptin, 1μg/mL PepstatinA) |
|
| Methods |
|
1. |
|
| 1) |
Plate HEK 293 cells on a 3.5 cm diameter dish. |
Comment 1
|

|
| 2) |
Approximately 24 h after plating, transfect plasmids. |
Comment 1
|

|
| 3) |
Approximately 24 h after transfection, remove medium and wash with prewarmed PBS (Dulbecco’s phosphate-buffered saline). |
Comment 0
|

|
| 4) |
Add 500 μL of medium lacking methionine and cysteine supplemented with 10% dialyzed FBS. |
Comment 1
|

|
| 5) |
Remove medium and add 500 μL of fresh medium as in 4. |
Comment 0
|

|
| 6) |
Add [35S]methionine or [35S] Protein Labeling Mixture. |
Comment 0
|

|
| 7) |
Incubate the cells in the CO2 incubator for the period required. |
Comment 0
|

|
| 8) |
Harvest cells as described in 2. For pulse-chase analysis, remove the medium and add normal growth medium, and then incubate the cells in the CO2 incubator. |
Comment 1
|
|
|
|
2. |
Coimmunoprecipitation assay |
| 1) |
Remove the medium and wash the cells twice with PBS. |
Comment 0
|

|
| 2) |
Add 1% NP-40 lysis buffer supplemented with protease inhibitors and incubate on ice for 20 min. |
Comment 0
|

|
| 3) |
Collect cell lysate using rubber policeman, and transfer into a microtube. |
Comment 0
|

|
| 4) |
Centrifuge at 13,000 × g at 4ºC for 20 min. |
Comment 0
|

|
| 5) |
Transfer the supernatant to a new microtube. |
Comment 0
|

|
| 6) |
Add equal volume of glycerol and mix by inversion. |
Comment 1
|

|
| 7) |
Aliquot and add antibodies to each tube. |
Comment 0
|

|
| 8) |
Stand still at 4ºC for one hour to overnight. |
Comment 1
|

|
| 9) |
Add 20 μL bed volume of Protein A Sepharose or Protein G Sepharose and 500 μL of lysis buffer, and rotate at 4ºC for 90 min. |
Comment 1
|

|
| 10) |
Centrifuge at 1,500 × g at 4ºC for 3 min, then remove supernatant. |
Comment 0
|

|
| 11) |
Add 700 μL of high ionic extraction buffer, and invert 30 times to mix. |
Comment 0
|

|
| 12) |
Centrifuge at to 1,500 × g at 4ºC for 3 min, then remove supernatant. |
Comment 0
|

|
| 14) |
Add to 700 μL of 10mM Tris-HCl, pH 7.5, and invert 30 times to mix. |
Comment 1
|

|
| 15) |
Centrifuge at 1,500 × g at 4ºC for 3 min, then remove supernatant |
Comment 0
|

|
| 16) |
Add 2 × Laemmli’s buffer and DTT, and incubate at 65ºC for 15 min. |
Comment 0
|

|
| 17) |
Centrifuge at 13,000 × g at room temperature for 10 min. |
Comment 0
|

|
| 18) |
Apply the supernatant to the gel for SDS-PAGE. |
Comment 0
|

|
| 19) |
Dry the gel after electrophoresis. |
Comment 0
|

|
| 20) |
Expose the gel to a phospho-imaging plate. |
Comment 0
|

|
| 21) |
Detect the radioactivity using phospho-image analyzer. |
Comment 0
|
|
|
| Figure & Legends |
Figure & Legends 
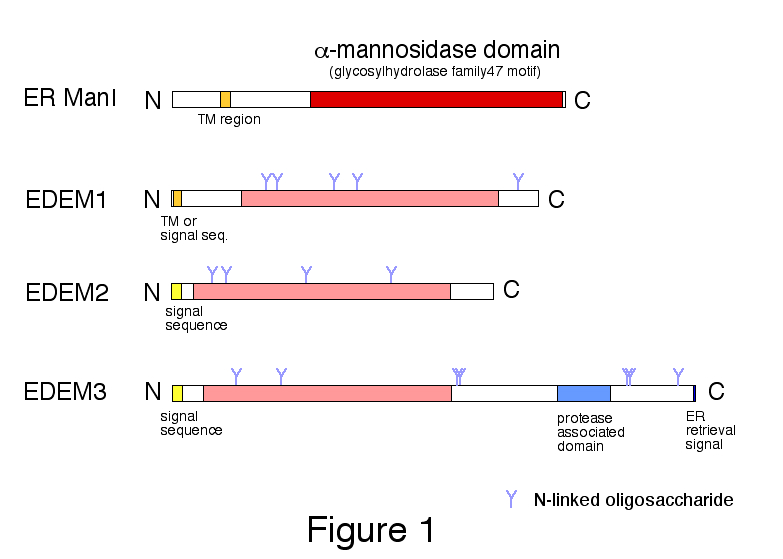
Fig. 1. Domain organization of human ER α-mannosidase I (ER ManI) and EDEMs.
Class I α1,2-mannosidase domain (glycosylhydrolase family 47 motif) is shown in red for ER ManI and in pink for EDEMs.
Copyright 2011 From "Animal Lectins: A Functional View" by G. R. Vasta and H. Ahmed. Modified by permission of Taylor and Francis Group, LLC, a division of Informa plc.

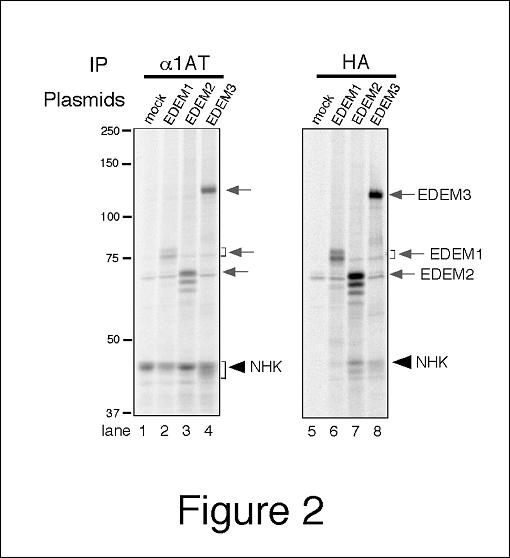
Fig. 2. Coimmunoprecipitation of EDEM1, 2, and 3 with ERAD substrate NHK.
HEK 293 cells were transfected with NHK (α1-antitrypsin null Hong Kong) and HA-tagged EDEM1, 2, or 3. Cells were treated with protease inhibitor MG132 for 3 h prior to metabolic labeling, and labeled with [35S] Protein Labeling Mixture for 1 h. Aliquot of the cell lysate is subjected to immunoprecipitation with anti-α1-antitrypsin antibody (lanes 1–4) or with anti-HA tag antibody (lanes 5–8). |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2014-09-08 16:57:52 |
- Clerc, S., Hirsch, C., Oggier, D. M., Deprez, P., Jakob, C., Sommer, T., and Aebi, M. (2009). Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J Cell Biol. 184, 159–72 [PMID : 19124653]
- Hirao, K., Natsuka, Y., Tamura, T., Wada, I., Morito, D., Natsuka, S., Romero, P., Sleno, B., Tremblay, L. O., Herscovics, A., Nagata, K., and Hosokawa, N. (2006). EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J Biol Chem. 281, 9650–8 [PMID : 16431915]
- Hosokawa, N., and Nagata, K. (2009). M-Type Lectins as Novel Components of Secretory Pathways. In "Animal Lectins: A Functional View" (G. R. Vasta and H. Ahmed, Eds.), pp. 165–169. CRC Press [PMID : not available]
- Hosokawa, N., Tremblay, L. O., Sleno, B., Kamiya, Y., Wada, I., Nagata, K., Kato, K., and Herscovics, A. (2010). EDEM1 accelerates the trimming of alpha1,2-linked mannose on the C branch of N-glycans. Glycobiology 20, 567–75 [PMID : 20065073]
- Hosokawa, N., Wada, I., Hasegawa, K., Yorihuzi, T., Tremblay, L. O., Herscovics, A., and Nagata, K. (2001). A novel ER alpha-mannosidase-like protein accelerates ER-associated degradation. EMBO Rep. 2, 415–22 [PMID : 11375934]
- Jakob, C. A., Bodmer, D., Spirig, U., Battig, P., Marcil, A., Dignard, D., Bergeron, J. J., Thomas, D. Y., and Aebi, M. (2001). Htm1p, a mannosidase-like protein, is involved in glycoprotein degradation in yeast. EMBO Rep. 2, 423–30 [PMID : 11375935]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Hosokawa, Nobuko,
(2014). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.27,4,2024 .
How to Cite this Work in Website:
Hosokawa, Nobuko,
(2014).
Isolation and binding assay of EDEMs.
Retrieved 27,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t98.
html source
Hosokawa, Nobuko,
(2014).
<b>Isolation and binding assay of EDEMs</b>.
Retrieved 4 27,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t98" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t98</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|