The hepatic asialoglycoprotein receptor was the first cellular receptor to be identified and isolated and the first lectin to be detected in mammals, which rapidly clears from blood circulation glycoproteins bearing glycan ligands with terminal galactose and N-acetylgalactosamine residues (Ashwell G, Morell AG. 1974). The smallest functional unit identifiable in aqueous solution possessed an estimated molecular weight of 500,000, which is consisting of two subunits with estimated molecular weights of 48,000 and 40,000, respectively (Hudgin LR et al.1974, Kawasaki T, Ashwell G. 1976) (see Fig. 3). The receptor is shown to modulate von Willebrand Factor homeostasis and is responsible for thrombocytopenia during systemic Streptococcus pneumoniae infection by eliminating platelets desialylated by the bacterium's neuraminidase (Grewal PK et al, 2008). In addition, this carbohydrate-mediated endocytosis is a key factor in the development and administration of glycoprotein pharmaceuticals. |
| Category | Sugar binding proteins |
| Protocol Name | Isolation and properties of a rabbit liver binding protein specific for asialoglycoproteins |
Authors
 |
Kawasaki, Toshisuke
Research Center for Glycobiotechnology, Ritsumeikan University
|
| KeyWords |
|
Reagents
 |
| ● |
|
| ● |
|
| ● |
Whatman GF/C discs (2.4 cm) |
| ● |
|
| ● |
Washing buffer: 0.1 M sodium acetate buffer, pH 6.0, containing 0.01 M EDTA, 0.2 M NaCl |
| ● |
Extraction buffer: 0.01 M Tris-HCl buffer, pH 7.8, containing 0.4 M KCl and1.0% Triton X100 |
| ● |
Equilibrium (Loading) buffer: 0.02 M Tris-HCl buffer, pH 7.8, containing 0.05 M CaCl2, 1.25 M NaCl and 0.5% Triton X-100 |
| ● |
Eluting buffer: 0.02 M ammonium acetate buffer, pH 6.0, containing 1.25 M NaCl and 0.5% Triton X-100 |
| ● |
Assay mixture: 0.05 M Tris-HCl buffer, pH 7.8, containing 1.0 M NaCl, 0.05 M CaC12, 0.1% Triton X-100 and 0.6% bovine serum albumin |
| ● |
Asialo-orosomucoid: prepared from orosomucoid (human alpha-1-acid glycoprotein) (Sigma-Aldrich, St. Louis, MO) by desialylation under mild acid treatment (in 0.05 M H2SO4 at 80ºC for 1 h) |
| ● |
1251-Asialo-orosomucoid (20 to 30 KBq per μg): prepared from asialo-orosomucoid by iodination with carrier free Na1251 using chloramine T (Sigma-Aldrich) |
| ● |
Asialo-orosomucoid affinity column: Sepharose 4B (50 mL), which had been activated with cyanogen bromide, was suspended in 100 mL of cold 0.1 M NaHCO3, pH 9.1. To this was added 5 mL of a 2% solution of asialo-orosomucoid and the coupling reaction was allowed to continue overnight on a rotary shaker at 4ºC |
|
Instruments
 |
| ● |
|
| ● |
|
| ● |
Multiwell filter plate vaccum manifold assembly for Whatman GF/C discs (2.4 cm) |
| ● |
Autogamma scintillation counter |
|
| Methods |
|
1. |
Acetone Powder Preparation |
| 1) |
The acetone and the Waring Blendor container were cooled to −20ºC. |
Comment 0
|

|
| 2) |
Fresh, or frozen and thawed rabbit liver was chilled on ice, minced well with scissors and blended with 10 volumes of cold acetone for two 30 sec periods. |
Comment 0
|

|
| 3) |
The suspension was immediately filtered under reduced pressure on Whatman No. 1 paper. As soon as the acetone disappeared from the surface, the cake was crumbled into the blender, extracted a second time, and filtered. |
Comment 0
|

|
| 4) |
The cake was washed with cold acetone, crumbled, and pushed quickly by hand through a fine wire window screen. |
Comment 0
|

|
| 5) |
The powder was allowed to air dry for 30 min, placed in a desiccator over NaOH, and stored under reduced pressure at 4ºC. |
Comment 0
|
|
|
|
2. |
Solubilization of Binding Protein |
| 1) |
Acetone powder was stirred for 30 min at 4ºC in 20 volumes of 0.1 M sodium acetate buffer, pH 6.0, containing 0.01 M EDTA and 0.2 M NaCl, and the suspension was centrifuged at 20,000 × g for 10 min. |
Comment 0
|

|
| 2) |
The supernatant fraction was discarded and the step was repeated twice, first with the same buffer and then with cold distilled water. |
Comment 0
|

|
| 3) |
The residual pellet was suspended by homogenization, or blending, in 20 volumes of extracting buffer consisting of 0.01 M Tris-HCl, pH 7.8, 0.4 M KCl and 1% Triton X-100. |
Comment 0
|

|
| 4) |
After stirring for 30 min at 4ºC, the debris was removed by centrifugation, the crude extract was decanted, made 0.05 M in CaCl2, and the pH adjusted to 7.8 with 1 M Tris-HCl buffer, pH 7.8. |
Comment 0
|
|
|
|
3. |
Purification of Binding Protein by Affinity Chromatography |
| 1) |
After equilibration of the asialo-orosomucoid affinity column with a loading buffer consisting of 0.01 M Tris-HCl, pH 7.8, 0.05 M CaCl2, 1.25 M NaCl and 0.5% Triton X-100, the crude extract was added in a ratio of 20 volumes of extract to 1 volume of resin. |
Comment 0
|

|
| 2) |
After washing the column several additional bed volumes of this buffer, the binding protein was eluted with a solution consisting of 0.02 M ammonium acetate buffer, pH 6.0, containing 1.25 M NaCl and 0.5% Triton X-100. |
Comment 0
|

|
| 3) |
The major portion of the binding activity was recovered in the 2nd bed volume of eluting buffer. |
Comment 0
|

|
| 4) |
Upon readjustment of the pH to 7.8 and addition of CaCl2 to 0.05 M, this material was adsorbed onto a second, smaller affinity column containing 1 mL of resin for each 3 mg of protein present. |
Comment 0
|

|
| 5) |
The binding protein, eluted with 0.02 M ammonium acetate buffer, pH 6.0, containing 0.5 M NaCl and 0.5% Triton X-100, was precipitated with an equal volume of cold (−70ºC) ethanol. The precipitate was washed once with 50% cold ethanol and dissolved in 2 mM Tris-HCl buffer, pH 7.8 (binding protein in water) or 0.1% Triton X-100 in 2 mM Tris-HCl buffer, pH 7.8 (binding protein in Triton X-100). |
Comment 0
|
|
|
|
4. |
|
| 1) |
The assay was carried out in disposable glass tubes, 10 × 75 mm. |
Comment 0
|

|
| 2) |
The incubation mixture contained, in a final volume of 0.50 mL, the following components: 0.05 M Tris-HCl buffer, pH 7.8, 1.0 M NaCl, 0.05 M CaC12, 0.1% Triton X-100, 0.6% bovine serum albumin, 1 μg 1251-asialo-orosomucoid (20 to 30 KBq per μg), 1 to 4 mg purified binding protein (or an equivalent amount of crude extract). |
Comment 0
|

|
| 3) |
The complete mixture was incubated at 25ºC for 30 min and then chilled in ice. |
Comment 0
|

|
| 4) |
The labeled complex was precipitated by the addition of 0.5 mL of a cold, saturated solution of ammonium sulfate, adjusted to pH 7.8 with solid Tris. |
Comment 0
|

|
| 5) |
After 10 min at 4ºC, the suspension was filtered on Whatman GF/C discs (2.4 cm) under reduced pressure and the incubation tube was washed three times with 1.0 mL of 0.4 saturated ammonium sulfate, pH 7.8, containing 10 mM CaCl2 and 0.1% bovine serum albumin. The filter was washed with an additional 7 mL of the same ammonium sulfate solution. |
Comment 0
|

|
| 6) |
The disc was removed and counted in a Autogamma scintillation counter. |
Comment 0
|
|
|
| Notes |
- Acetone Powder Preparation: Approximately 25 to 30 g of acetone powder were recovered from 100 g (wet weight) of rabbit liver. Although the binding activity of the powder remained stable for several weeks, better recoveries were obtained when the powder was extracted within a few days after preparation.
- Purification of Binding Protein by Affinity Chromatography: From 50 g of acetone powder, 10 to 15 mg of final product were routinely recovered. This material is stored at 4ºC with complete retention of activity for several months.
- Binding Assay: Appropriate blanks were prepared by incubation of the binding protein without added ligand, precipitation as above, and addition of 1 mg of 1251-asialo-orosomucoid immediately before filtration. The values so obtained, approximately 0.2% of the total radioactivity added, were subtracted from each sample. Under these conditions, the assay was linear and proportional to the amount of binding protein present in the incubation.
|
| Figure & Legends |
Figure & Legends 
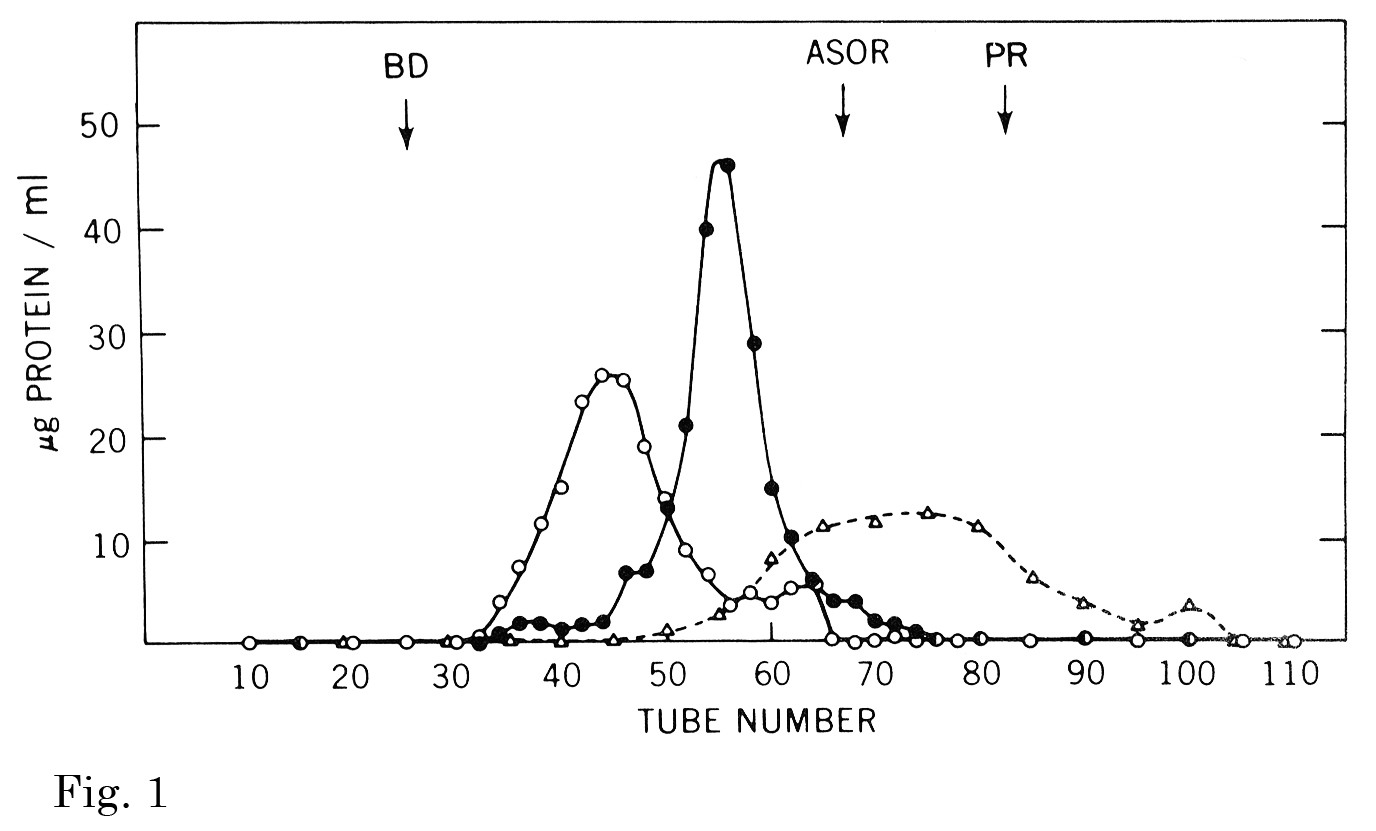
Fig. 1. The effect of calcium and ligand on the pattern of Sepharose 4B gel filtration of the binding protein.
Three separate incubations (1-3) of the binding protein were subjected to gel filtration on the same column of Sepharose 4B (1.5 × 116 cm). (1) the binding protein (1.2 mg) was incubated with 1251-asialo-orosomucoid (970 μg, 2.6 × 106 cpm) in an assay mixture except that bovine serum albumin was omitted. After incubation at room temperature for 20 min, the solution was charged onto the column and eluted with the same buffer except that the NaCl and CaCl2 concentration was reduced to 0.5 M and 5 mM, respectively. The elution pattern of the bound complex (○) represents the protein content of the binding protein. (2) binding protein (1.5 mg) was incubated and chromatographed as in (1) except that calcium was omitted from the elution buffer. (3) binding protein (1.4 mg) was incubated and chromatographed as in (1) except that the radioactive ligand was omitted. The symbols are: ○, complete system; ●, minus calcium; △, minus 1251-asialo-orosomucoid.
This figure was originally published in J Biol Chem. Kawasaki T. et al. “Chemical and physical properties of an hepatic membrane protein that specifically binds asialoglycoproteins” 1976, 251(5):1296–302. © the American Society for Biochemistry and Molecular Biology.

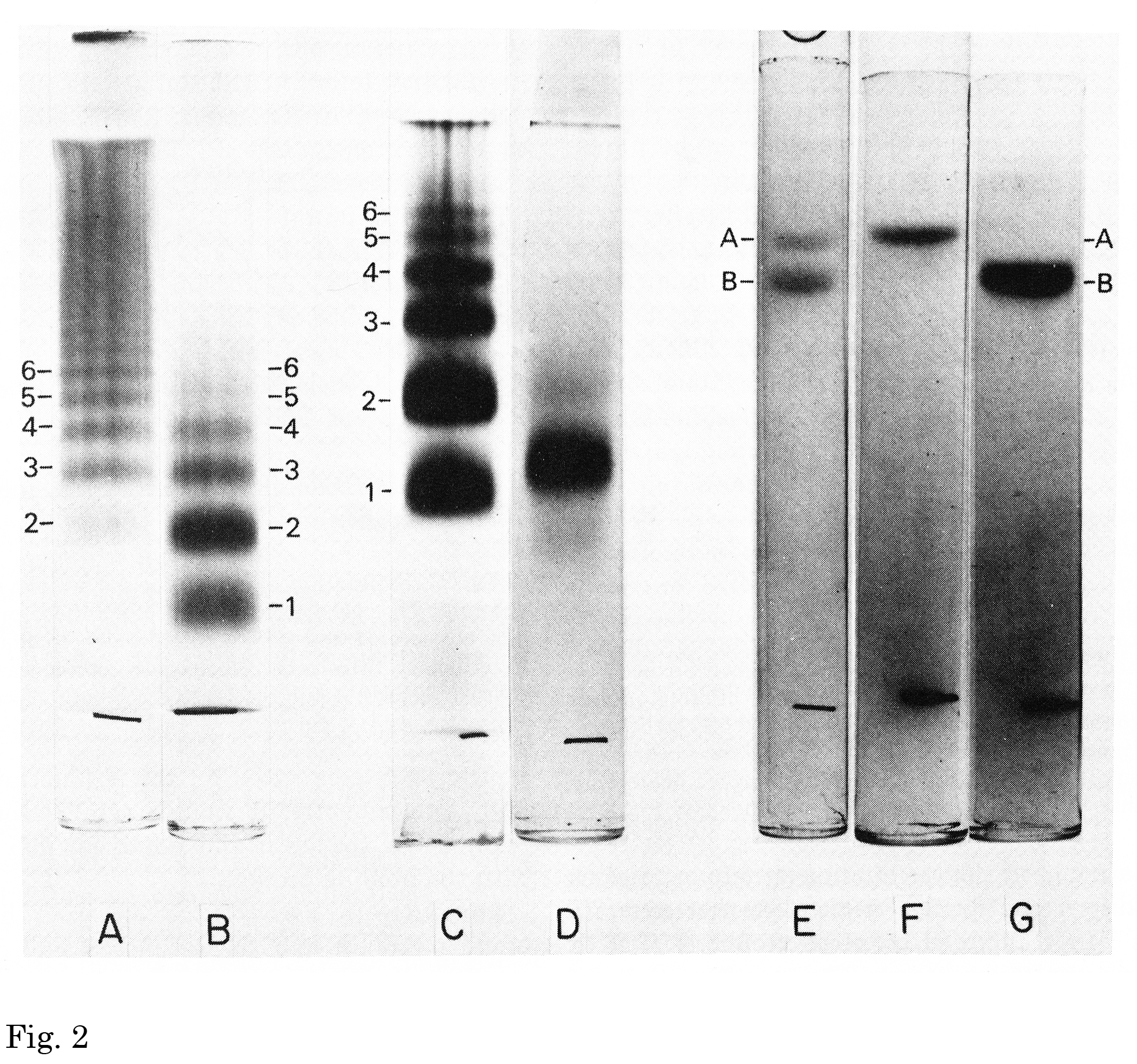
Fig. 2. Disc acrylamide gel electrophoresis of the binding protein and its subunits.
A, an aqueous solution of the binding protein (30 μg) was applied onto a 3% acrylamide gel. Electrophoresis was carried out using a multiphasic buffer system “System A” (pH 9.45) (Rodbard D, Chrambach A, 1971) for 2.5 h at 4ºC. Protein bands were stained with Coomassie brilliant blue G-250 in 12.5% trichloroacetic acid. B, a 0.1% Triton X-100 solution of the binding protein was applied on to a 3% gel. C, a 0.1% Triton X-100 solution of the binding protein was applied to a 4% gel. D, a 0.1% Triton X-100 solution of the binding protein made 1.0% in Triton X-100 and stored at room temperature for 1 h, was applied to a 4% gel containing 0.1% Triton X-100. E, an aqueous solution of the binding protein was incubated in 1.0% sodium dodecyl sulfate (SDS)/0.5 mM EDTA/0.01 M sodium phosphate, pH 7.0, for 3 days at 48ºC. The solution was then made 10% in β-mercaptoethanol and 4 M in urea. Incubation was continued for an additional 3 h at 48ºC before applying the sample to a 10% gel containing 0.1% SDS. Electrophoresis was carried out at room temperature for 6 h according to the method of Weber and Osborn (1969). F, subunit A (7 μg) was incubated in 1% SDS, 0.01 M sodium phosphate, pH 7.0 for 24 h. The sample was made 10% in β-mercaptoethanol, incubated 1 additional hour at 48ºC and applied to a 10% gel containing 0.1% SDS. Electrophoresis was carried out as in E. G, subunit B (14 μg) was incubated and analyzed by electrophoresis as described in F.
This figure was originally published in J Biol Chem. Kawasaki T. et al. “Chemical and physical properties of an hepatic membrane protein that specifically binds asialoglycoproteins” 1976, 251(5):1296–302. © the American Society for Biochemistry and Molecular Biology.

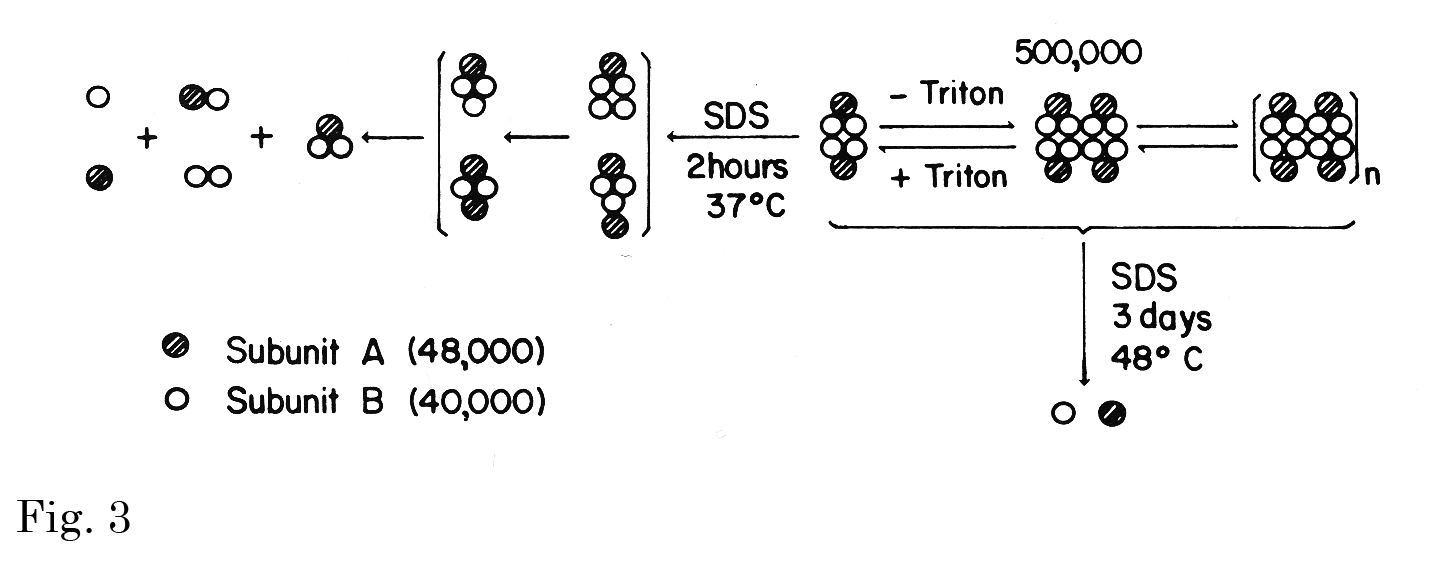
Fig. 3. Hypothetical formulation of subunit interaction in producing aggregation.
The molecular weights of two subunits (40,000 and 48,000) that of the smallest functional unit (500,000) were estimated by Ferguson plot analyses of SDS-PAGE data at four different acrylamide concentrations from 5 to 12.5% and those of PAGE data at seven different acrylamide concentrations from 3 to 6%, respectively. The species in brackets were observed only after suberimidate cross-linking.
This figure was originally published in J Biol Chem. Kawasaki T. et al. “Chemical and physical properties of an hepatic membrane protein that specifically binds asialoglycoproteins” 1976, 251(5):1296–-302. © the American Society for Biochemistry and Molecular Biology. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-02-25 13:36:07 |
- Ashwell, G., and Morell, A.G. (1974) The role of surface carbohydrates in the hepatic recognition and transport of circulating glycoproteins. Adv Enzymol. 41, 99–128 [PMID : 4609051]
- Hudgin, R.L., Pricer, W.E. JR, Ashwell, G., Stockert, R.J., and Morell, A.G. (1974) The isolation and properties of a rabbit liver binding protein specific for asialoglycoproteins. J Biol Chem. 249, 5536–5543 [PMID : 4370480]
- Kawasaki, T., and Ashwell, G. (1976) Chemical and physical properties of an hepatic membrane protein that specifically binds asialoglycoproteins. J Biol Chem. 251, 1296–1302 [PMID : 1254568]
- Grewal, P.K., Uchiyama, S., Ditto, D., Varki, N., Le, D.T., Nizet, V., and Marth, J.D. (2008) The Ashwell receptor mitigates the lethal coagulopathy of sepsis. Nat Medicine 14, 648–655 [PMID : 18488037]
- Weber, K., and Osborn, M. (1969) The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 244, 4406–4412 [PMID : 5806584]
- Rodbard, D., and Chrambach, A. (1971) Estimation of molecular radius, free mobility, and valence using polyacylamide gel electrophoresis. Anal Biochem. 40, 95–134 [PMID : 5550151]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Kawasaki, Toshisuke,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.25,4,2024 .
How to Cite this Work in Website:
Kawasaki, Toshisuke,
(2015).
Isolation and properties of a rabbit liver binding protein specific for asialoglycoproteins.
Retrieved 25,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t97.
html source
Kawasaki, Toshisuke,
(2015).
<b>Isolation and properties of a rabbit liver binding protein specific for asialoglycoproteins</b>.
Retrieved 4 25,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t97" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t97</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|