The measurement of intracellular nucleotide sugar is one of the key technologys in glyobiology fields. Since we have been engaged in the development of yeast strains, which can produce recombinant glyocoproteins with heterologous sugar chains, it is essential to generate new nucleotide sugars from endogenous nucleotide sugars through the expression of foreign genes responsible for the conversion of nucleotide sugars and to measure the amounts of produced nucleotide sugars inside the cells.
Here we describe the methods on the extraction of intracellular nucleotide sugars with formic acid saturated with 1-butanol and its analysis by HPLC system. |
| Category | Nucleotide sugar transporters |
| Protocol Name | Analysis of intracellular nucleotide sugars |
Authors
 |
Oka, Takuji
Faculty of Biotechnology and Life Science, Department of Applied Microbial Technology, Sojo University
Jigami, Yoshifumi
*
Research Center for Medical Glycoscience, National Institute of Advanced Industrial Science and Technology (AIST)
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
Extraction buffer: 1 M formic acid saturated with 1-butanol |
| ● |
20 mm triethylamine acetate (TEAA) buffer (pH 7.0) |
| ● |
(Option) solvent A (20 mM K2HPO4-KHPO4, pH 7.5) |
| ● |
(Option) solvent B (200 mM K2HPO4-KHPO4, pH 7.5) |
|
Instruments
 |
| ● |
DISMIC-03CP (Advantec Co., Ltd., Ehime, Japan) |
| ● |
Cosmosil 5C18-AR-II column (Nacalai Tesque Inc., Kyoto, Japan) |
| ● |
Develosil RPAQUEOUS column (250 × 4.6 mm, Nomura Chemical Co., Ltd., Seto, Japan) |
| ● |
HPLC system equipped with UV detector (LC-10, Shimadzu Corporation, Kyoto, Japan) |
| ● |
ESI-MS system (Bruker Daltonik GmbH, Bremen, Germany) |
| ● |
Centrifuge concentrator (TAITEC Co., Ltd., Koshigaya, Japan) |
| ● |
(Option) CarboPac PA1 anion-exchange column (250 × 4.0 mm, Dionex Corp., Sunnyvale, CA) |
|
| Methods |
|
1. |
Extraction of intracellular sugar nucleotide |
| 1) |
Cultivation and harvest of cells.
Yeast cells (OD600 nm = 50) or Mammalian cells (8 × 107 cells) |
Comment 0
|

|
| 2) |
Add 5 mL of ice-cold 1 mL formic acid saturated with 1-butanol to the cells. |
Comment 0
|

|
| 4) |
Centrifuge at 13,000 × g for 5 min to remove cell debris. |
Comment 0
|

|
| 5) |
Dry the supernatant by centrifuge concentrator. |
Comment 0
|

|
| 7) |
Filtrate a membrane filter with a pore size of 0.45 μm (DISMIC-03CP). |
Comment 0
|
|
|
|
2. |
HPLC analysis of intracellular sugar nucleotide |
| 1) |
Analyze the above extracted intracellular sugar nucleotide (40 μL) by HPLC (LC-10) on a cosmosil 5C18-AR-II column. The column is equilibrated with 20 mM TEAA buffer (pH 7.0) at a flow rate of 1 mL·min−1. UDP-sugars are detected by UV260 nm absorbance. |
Comment 0
|

|
| 2) |
The peaks of detected target sugar nucleotides are collected based on the retention times of the standards. |
Comment 0
|

|
| 3) |
The collected sugar nucleotides are dried by centrifuge concentrator and resolved in 40 μL of distilled water. |
Comment 0
|

|
| 4) |
The structures of the UDP-sugars are analyzed by ESI-MS to determine its molecular mass. Mass spectra are acquired on an Esquire 3,000-plus instrument in the negative-ion mode. Conditions for ESI-MS are as follows: 68.95 kPa nebulizer flow, 300°C nozzle temperature, and 5.0 L·min−1 flow of drying gas (N2). |
Comment 0
|

|
| 5) |
The collected sugar nucleotides are reseparated on the HPLC system (LC-10) with a Develosil RPAQUEOUS column (250 × 4.6 mm). The column is equilibrated with 20 mM TEAA buffer (pH 7.0) at a flow rate of 1 mL·min−1. UDP-sugars are detected by UV260 nm absorbance.
(Option)
Alternatively, the collected sugar nucleotides can be separated on the HPLC system (LC-10,) with a CarboPac PA1 anion-exchange column. The column was equilibrated with solvent A (20 mM K2HPO4-KHPO4, pH 7.5) at a flow rate of 0.7 mL/min for 5 min, and analyzed isocratically with solvent B (200 mM K2HPO4-KHPO4, pH 7.5) at a flow rate of 0.7 mL/min for 30 min. |
Comment 0
|
|
|
| Notes | If target sugar nucleotides cannot be separated with a reverse phase column, we recommend the use of anion-exchange column to separate target sugar nucleotides as described in “Option”. |
| Figure & Legends |
Figure & Legends 
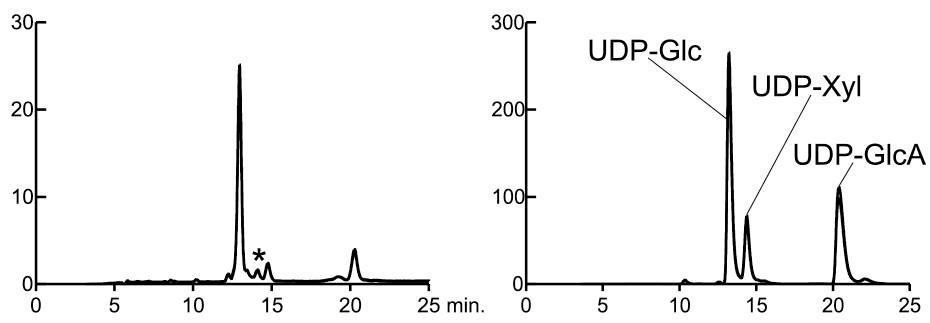
Fig. 1. UDP-Glc, UDP-GlcA, and UDP-Xyl content of CHO cells.
Nucleotide sugars of wild-type cells were separated in a two-step HPLC (left panel). Fractions of the first HPLC run were collected, and combined fractions of UDP-Glc, UDP-GlcA, and UDP-Xyl (based on the position of standards (right panel)) were applied to a second run using Cosmosil 5C18-AR-II column. *location of UDP-Xyl as confirmed by mass spectrometry.
This figure was originally published in J Biol Chem. Bakker H, Oka T. et al. "Functional UDP-xylose Transport across the Endoplasmic Reticulum/Golgi Membrane in a Chinese Hamster Ovary Cell Mutant Defective in UDP-xylose Synthase" 2009 284:2576-583. © the American Society for Biochemistry and Molecular Biology.
|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2014-05-28 09:11:26 |
- Oka, T., and Jigami, Y. (2006) Reconstruction of de novo pathway for synthesis of UDP-glucuronic acid and UDP-xylose from intrinsic UDP-glucose in Saccharomyces cerevisiae. FEBS J. 273, 2645–2657. [PMID : 16817893]
- Oka, T., Nemoto, T., and Jigami, Y. (2007) Functional analysis of Arabidopsis thaliana RHM2/MUM4, a multidomain protein involved in UDP-D-glucose to UDP-L-rhamnose conversion. J Biol Chem. 282, 5389–5403. [PMID : 17190829]
- Bakker, H., Oka, T., Ashikov, A., Yadav, A., Berger, M., Rana, N.A., Bai, X., Jigami, Y., Haltiwanger, R.S., Esko, J.D., and Gerardy-Schahn, R. Functional UDP-xylose transport across the endoplasmic reticulum/Golgi membrane in a Chinese hamster ovary cell mutant defective in UDP-xylose Synthase. J Biol Chem. 284, 2576–2583. [PMID : 19028698]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Oka, Takuji,
Jigami, Yoshifumi,
(2014). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.25,4,2024 .
How to Cite this Work in Website:
Oka, Takuji,
Jigami, Yoshifumi,
(2014).
Analysis of intracellular nucleotide sugars.
Retrieved 25,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t96.
html source
Oka, Takuji,
Jigami, Yoshifumi,
(2014).
<b>Analysis of intracellular nucleotide sugars</b>.
Retrieved 4 25,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t96" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t96</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|