N-Acetylglucosamine-6-O-sulfotransferase (GlcNAc6ST) transfers a sulfate group from 3’-phosphoadenosine 5’-phosphosulfate (PAPS) to an N-acetylglucosamine (GlcNAc) residue, which is usually located at the non-reducing end of glycans. Five members of the GlcNAc6ST family have been cloned in humans, four of which have orthologs in mice. Among the GlcNAc6STs, GlcNAc6ST-1 and GlcNAc6ST-2 have been confirmed to be involved in the synthesis of sialyl 6-sulfo Lewis X, which is a major class of L-selectin ligands in lymphocyte homing. GlcNAc-6-sulfation mainly regulates the rolling velocity of lymphocytes in high endothelial venules of peripheral lymph nodes. In this section, we describe an in vitro assay for GlcNAc6STs. GlcNAc6STs fused with Protein A are expressed in mammalian cell lines and then purified from the culture media. Various oligosaccharides derived from N-linked or O-linked glycans are applicable to the assay. |
| Category | Glycosyltransferases & related proteins |
| Protocol Name | Enzyme assay of N-acetylglucosamine-6-sulfotransferases for selectin ligands |
Authors
 |
Uchimura, Kenji
Nagoya University Graduate School of Medicine, Department of Biochemistry
|
| KeyWords |
|
Reagents
 |
| ● |
Phosphate buffered saline (PBS) |
| ● |
Dulbecco’s modified Eagle’s medium (DMEM: Invitrogen/Life Technologies, Carlsbad, CA) |
| ● |
Fetal bovine serum (FBS: Invitrogen/Life Technologies) |
| ● |
LipofectAMINE PLUS (Invitrogen/Life Technologies) |
| ● |
IgG-Sepharose (GE Healthcare, Little Chalfont, UK) |
| ● |
|
| ● |
Thin-layer chromatography (TLC) plates (Merck Millipore, Billerica, MA) |
| ● |
[35S] PAPS (1.9 Ci/mmol: PerkinElmer, Waltham, MA) |
|
Instruments
 |
| ● |
Water bath incubator, 37°C |
| ● |
|
| ● |
BAS2000 bioimaging analyzer (Fujifilm, Tokyo, Japan) |
|
| Methods |
|
1. |
Expression and Purification of GlcNAc6STs Fused with Protein A |
| 1) |
Construction of the plasmids encoded fusion proteins of GlcNAc6STs to the IgM signal sequence and protein A in the N-terminal region are as described1). |
Comment 0
|

|
| 2) |
Culture COS-7 cells in DMEM containing 10% FBS on a 10-cm dish and then transfect 4 μg of a GlcNAc6ST expression plasmid using LipofectAMINE PLUS according to manufacturer instructions. |
Comment 0
|

|
| 3) |
Replace the medium with DMEM containing 2% IgG-free FBS after 24 h. Culture cells for another 48 h. |
Comment 0
|

|
| 4) |
Collect the culture medium and then mix with IgG-Sepharose (10 μL of resin/10 mL of culture). |
Comment 0
|

|
| 5) |
Incubate the mixture at 4°C for 3 h with gently rocking to adsorb the protein A fusion GlcNAc6STs expressed in the medium to the resin. |
Comment 0
|

|
| 6) |
Collect the resin by centrifugation and wash three times with PBS. |
Comment 0
|

|
| 7) |
Suspend the resin in 30 μL of 50 mM Tris-HCl, pH 7.5, and used as enzymes. |
Comment 0
|
|
|
|
2. |
Assay of GlcNAc6ST activities toward oligosaccharides |
| 1) |
The standard reaction mix in a 1.5 mL tube contains 1 μmol Tris-HCl, pH 7.5, 0.2 μmol MnCl2, 0.04 μmol adenosine 5’-monophosphate, 2 μmol sodium fluoride, 20 nmol oligosaccharides, 150 pmol [35S]-PAPS (1.5 × 106 cpm), 0.05% of Triton-X and 1 μL of fusion protein suspension in a final volume of 20 μL. |
Comment 0
|

|
| 2) |
Incubate the tube containing the reaction mix at 30°C for 1 h. |
Comment 0
|

|
| 3) |
Apply aliquots of 2 μL of the reaction mix to TLC plates. The plates are pre-coated with cellulose 0.1-mm thick. |
Comment 0
|

|
| 4) |
Develop the plates with a developing solvent, ethanol/pyridine/n-butyl alcohol/water/acetic acid (100:10:10:30:3, by volume). |
Comment 0
|

|
| 5) |
End the development when the solvent front reaches to the top of the plates. The 35S-labeled oligosaccharides migrate faster than [35S]-PAPS. |
Comment 0
|

|
| 6) |
Visualize and measure the radioactivity of the 35S-labeled oligosaccharides with a BAS2000 bioimaging analyzer. |
Comment 0
|
|
|
| Notes |
- Oligosaccharide substrates tested are GlcNAcβ1-3Galβ1-4GlcNAc, GlcNAcβ1-6Man-O-methyl, GlcNAcβ1-2Man, GlcNAcβ1-6[Galβ1-3]GalNAc-p-nitrophenyl (Core2-pNP) and GlcNAcβ1-3GalNAc-p-nitrophenyl (Core3-pNP)1).
- TLC is suitable to handle a large number of samples.
|
| Figure & Legends |
Figure & Legends 
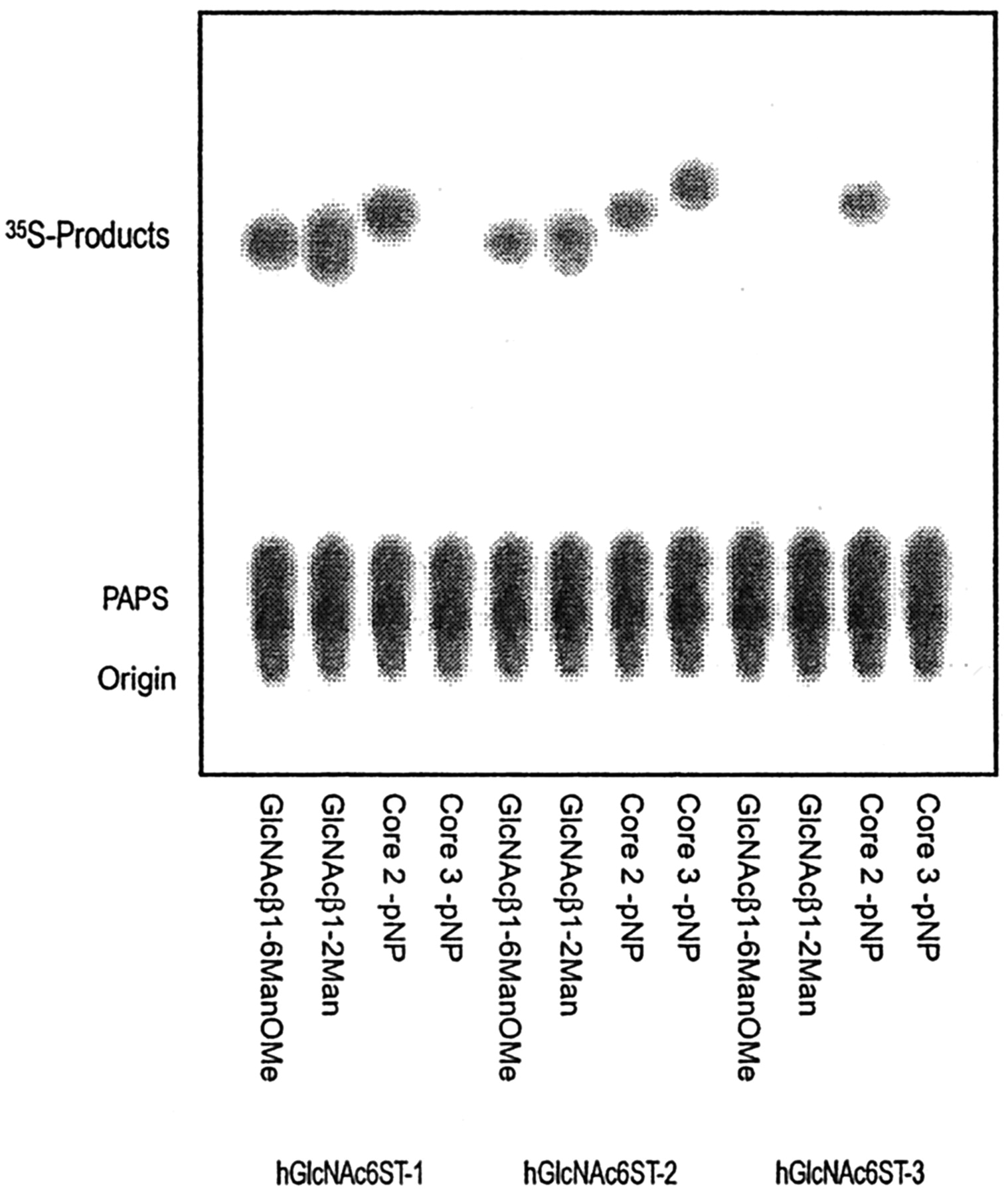
Fig. 1. Sulfation of various oligosaccharides by human GlcNAc6ST-1, -2 , and -3.
Various oligosaccharides were incubated with the protein A-fused GlcNAc6ST-1, -2, and -3 under standard assay conditions. Aliquots of 2 μL of each reaction were applied to a TLC plate and then developed with ethanol-pyridine-n-butyl alcohol-water-acetate (100:10:10:30:3 (v/v), respectively). 35S-Labeled products were visualized, and the radioactivity was measured with a BAS2000 bioimaging analyzer.
This figure was originally published in J Biol Chem. Uchimura K. et al. "Specificities of N-acetylglucosamine-6-O-sulfotransferases in relation to L-selectin ligand synthesis and tumor-associated enzyme expression" 2002, 277(6):3979–84. © the American Society for Biochemistry and Molecular Biology. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-10-01 13:37:50 |
- Uchimura, K., El-Fasakhany, F.M., Hori, M., Hemmerich, S., Blink, S.E., Kansas, G.S., Kanamori, A., Kumamoto, K., Kannagi, R., and Muramatsu, T. (2002) Specificities of N-acetylglucosamine-6-O-sulfotransferases in relation to L-selectin ligand synthesis and tumor-associated enzyme expression. J Biol Chem. 277, 3979–3984 [PMID : 11726653]
- Uchimura, K., Gauguet, J.M., Singer, M.S., Tsay, D., Kannagi, R., Muramatsu, T., von Andrian, U.H., and Rosen, S.D. (2005) A major class of L-selectin ligands is eliminated in mice deficient in two sulfotransferases expressed in high endothelial venules. Nat Immunol. 6, 1105–1113 [PMID : 16227986]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Uchimura, Kenji,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.26,4,2024 .
How to Cite this Work in Website:
Uchimura, Kenji,
(2015).
Enzyme assay of N-acetylglucosamine-6-sulfotransferases for selectin ligands.
Retrieved 26,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t85.
html source
Uchimura, Kenji,
(2015).
<b>Enzyme assay of <em>N</em>-acetylglucosamine-6-sulfotransferases for selectin ligands</b>.
Retrieved 4 26,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t85" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t85</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|