Glycosylphosphatidylinositol (GPI) anchoring of proteins is a conserved post-translational modification in the endoplasmic reticulum (ER). Once GPI is transferred to proteins, it is transported to the cell surface via the Golgi apparatus. A GPI-anchored protein, decay accelerating factor (DAF, CD55), acts as a complement regulatory protein to protect host cells from injury by complement activation. DAF is useful for monitoring transport of GPI-anchored proteins from the ER to the Golgi apparatus, because its molecular size at the ER is clearly separated from that of its mature counterpart that has reached the Golgi. The ER form (~45 kDa) of DAF is transported to the Golgi where modification of its O-glycans and N-glycan results in the mature form (70–75 kDa). Here, we report a protocol for pulse-chase analysis of DAF. |
| Category | GPI anchored proteins |
| Protocol Name | Transport of GPI anchored proteins from ER to Golgi ~Pulse-chase transport analysis of DAF from the ER to the Golgi |
Authors
 |
Fujita, Morihisa
Research Institute for Microbial Diseases, Osaka University
Maeda, Yusuke
Research Institute for Microbial Diseases, Osaka University
Kinoshita, Taroh
*
Research Institute for Microbial Diseases, Osaka University
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
Cells highly expressing human DAF |
| ● |
|
| ● |
Phosphate-buffered saline (PBS) |
| ● |
|
| ● |
DMEM without L-methionine and L-cysteine (Gibco/Life Technologies, Carlsbad, CA 21013-024) |
| ● |
[35S]-methionine/cysteine (example: EXPRE35S35S Protein Labeling Mix (Perkin Elmer, Waltham, MA NEG772)) |
| ● |
Chasing cocktail stock (100×): 150 mM methionine and 120 mM cysteine
100 mM NaN3 |
| ● |
PBS containing 10 mM NaN3 |
| ● |
RIPA buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) |
| ● |
Mouse anti-human DAF (IA10) (BD, Franklin Lakes, NJ) |
| ● |
Protein-A-Sepharose (or Protein-G-Sepharose) |
| ● |
TNET buffer (100 mM Tris-HCl (pH 7.4), 100 mM NaCl, 5 mM EDTA, 1% Triton X-100) |
|
Instruments
 |
| ● |
Incubator (37˚C), centrifuge, rotator, fluorescent image analyzer (example: FLA-5100 (Fujifilm, Tokyo, Japan)) |
|
| Methods |
|
1. |
Pulse-chase analysis of DAF |
| 2) |
Centrifuge at 1400 rpm for 3min. |
Comment 0
|

|
| 3) |
Resuspend in 5.5 mL methionine- and cysteine-free DMEM / 10% FCS. |
Comment 0
|

|
| 5) |
Pulse with 55 μL (550 μCi) [35S]-methionine/cysteine for 10 min. |
Comment 0
|

|
| 6) |
Add 1/100 volume of chasing cocktail (55 μL). |
Comment 0
|

|
| 7) |
Take up 1 mL cell suspension into new tube containing 100 μL of 100 mM NaN3 at varying time points (0, 10, 20, 40, 60 min) and keep on ice. |
Comment 0
|

|
| 8) |
Wash with ice-cold PBS-containing 10 mM NaN3. |
Comment 0
|

|
| 9) |
Lyse cells in 0.6 mL of RIPA buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS) containing protease inhibitor cocktail for 30 min on ice. |
Comment 0
|

|
| 10) |
Centrifuge at 13,000 × g for 15 min at 4˚C. |
Comment 0
|

|
| 11) |
Aspirate supernatant into a fresh tube and rotate with 1.5 μg/tube monoclonal anti-DAF antibody (IA10) and 30 μL Protein-A-Sepharose.
(Protein-A-Sepharose should pre-incubate with non-radiolabeled cell lysate for 1 h before starting immunoprecipitation to remove non-specific binders). |
Comment 0
|

|
| 12) |
Wash Protein-A-Sepharose with 1 mL TNET buffer 4 times. |
Comment 0
|

|
| 13) |
SDS-PAGE under reducing conditions. |
Comment 1
|

|
| 14) |
Analyze using a BAS 1000 analyzer. |
Comment 0
|
|
|
| Figure & Legends |
Figure & Legends 
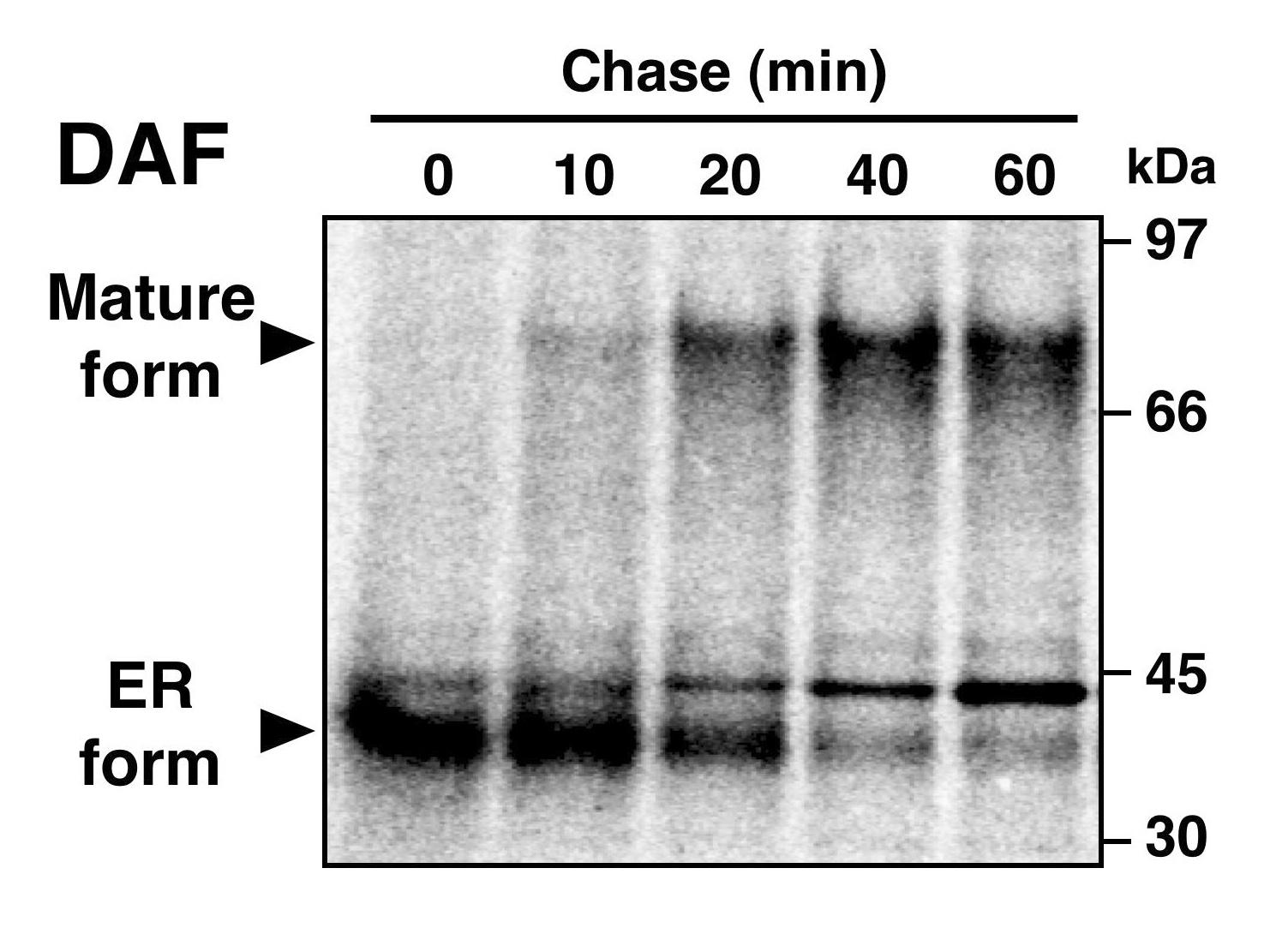
Pulse-chase of DAF.
CHO-K1 cells stably transfected with human DAF were pulse-labeled with a mixture of [35S]-methionine and -cysteine followed by chasing for the indicated times. DAF was immunoprecipitated using a monoclonal anti-DAF antibody, separated by SDS-PAGE and analyzed using a FLA-5100 analyzer. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-01-07 14:25:31 |
- Tanaka, S., Y. Maeda, Y. Tashima and T. Kinoshita, (2004) Inositol-deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279, 14256–14263 [PMID : 14734546]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Fujita, Morihisa,
Maeda, Yusuke,
Kinoshita, Taroh,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.25,4,2024 .
How to Cite this Work in Website:
Fujita, Morihisa,
Maeda, Yusuke,
Kinoshita, Taroh,
(2015).
Transport of GPI anchored proteins from ER to Golgi ~Pulse-chase transport analysis of DAF from the ER to the Golgi.
Retrieved 25,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t77.
html source
Fujita, Morihisa,
Maeda, Yusuke,
Kinoshita, Taroh,
(2015).
<b>Transport of GPI anchored proteins from ER to Golgi ~Pulse-chase transport analysis of DAF from the ER to the Golgi</b>.
Retrieved 4 25,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t77" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t77</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|