N-glycanase [peptide-N4-(N-acetyl-β-D-glucosaminyl) asparagine amidase, EC 3.5.1.52], also called peptide N-glycanase, PNGase, glycopeptidase and glycoamidase, is a kind of amidase that acts on the glycosylamide linkages between the innermost GlcNAc and asparagine residues of high mannose, hybrid, and complex (bi, tri-, or tetraantennary) oligosaccharide chains from N-linked glycoproteins (Fig.1). N-glycanase F (PNGase F) (Plummer TH Jr et al. 1984) from Flavobacterium meningosepticum is assumed to hydrolyze the glycosylamine linkage by a mechanism similar to that of the analogous almond enzyme (glycopeptidase A) (Takahashi N, Nishibe H. 1978), and the aspartylglycosylamine amidohydrolase (EC3.5.1.26), (Makino M et al. 1966), which was first found in blood serum and tissues cleaving the acid-amide linkage only in a small molecules consisting of one Asn linked to one GlcNAc residue. |
| Category | N-Glycans |
| Protocol Name | |
Authors
 |
Kawasaki, Nobuko
Research Center for Glycobiotechnology, Ritsumeikan University
|
| KeyWords |
|
Reagents
 |
| ● |
PNGase F (from Flavobacterium meningosepticum, molecular weight 35.5k) is commercially available from a number of sources. Enzymes purified from culture filtrate, New England BioLabs Inc., Ipswich, MA, Sigma-Aldrich, St. Louis, MO (proteomics grade) etc. Recombinant enzymes (expressed in E. coli) from Roche Diagnostics GmbH, Mannheim, Germany (N-glycosidase F), Sigma-Aldrich, Takara Bio Inc., Otsu, Japan etc. |
| ● |
Glycopeptidase A (from almond emulsion), also called glycoamidase A is available from Seikagaku Corp., Tokyo Japan, Sigma-Aldrich, and Roche Diagnotics GmbH. |
| ● |
|
| ● |
7.5% Nonidet P-40 (NP-40) |
| ● |
0.2 M sodium phosphate buffer, or 0.2 M Tris-HCl buffer, pH 7.5–8.5 |
| ● |
1% PMSF dissolved in 2-propyl alcohol |
| ● |
|
| ● |
|
| ● |
|
| ● |
50 mM–100mM ammonium bicarbonate buffer, pH 8.5 |
| ● |
C18 Sep-Pak cartridge (Waters Corp., Milford, MA) |
| ● |
pepsin (alternatively, trypsin and chymotrypsin) |
|
Instruments
 |
| ● |
Reaction Incubator or water bath (100°C, 37°C) |
|
| Methods |
|
1. |
In solution deglycosylation of N-linked glycoprotein with PNGase F. |
| 1) |
Heat a glycoprotein sample (10–50 μL, 1–2 mg/mL) in 0.2 M sodium phosphate buffer or 0.2 M Tris-HCl buffer, pH 7.5–8.5, containing 0.5% SDS, 50 mM 2-mercaptoethanol at 100°C for 3 min. |
Comment 1
|

|
| 2) |
Put 10 μL of the denatured glycoprotein sample solution into a microtube. |
Comment 1
|

|
| 3) |
Add 10 μL of 0.2 M sodium phosphate buffer or 0.2 M Tris-HCl buffer, pH 7.5, containing 2 mM EDTA and 0.03% PMSF. |
Comment 1
|

|
| 4) |
Add 5 μL of 7.5% NP-40 (the concentration of NP-40 is 7 fold excess that of SDS in the reaction mixture). |
Comment 1
|

|
| 5) |
Add 5 μL of PNGase F solution to give a final enzyme concentration of 0.5–10 U (IU) /mL. |
Comment 1
|

|
| 7) |
Stop the digestion by boiling the reaction mixture for 5 min. |
Comment 0
|

|
| 8) |
Analyze aliquots of the reaction product.⇒SDS-PAGE, Lectin-blotting, etc |
Comment 1
|

|
| 9) |
Add 3 volumes of ice-cold ethanol.⇒Centrifuge at 5,000 × g for 10 min⇒Save the supernatant to analyze the released oligosaccharides. |
Comment 1
|
|
|
|
2. |
Releasing the N-glycans from the glycopeptide with PNGase F (Kawasaki N et al, 2009). |
| 1) |
Dissolve glycopeptide(s) sample (~100 μg) in 200 μL of 50 mM ammonium bicarbonate buffer, pH 8.5. |
Comment 1
|

|
| 4) |
Separate the N-glycans from de-N-glycosylated peptides by passage through a reverse phase C18 Sep-Pak cartridge (1 mL). |
Comment 1
|

|
| 5) |
Wash the cartridge with 5% acetic acid. |
Comment 0
|

|
| 6) |
Elute the cartridge with 3 mL each of 20 and 40% propanol in 5% acetic acid (a third 60% propanol fraction can be collected but usually glycopeptides will elute earlier. The alternative water/acetonitrile solvent system can also be used). |
Comment 0
|

|
| 7) |
Pass through (N-glycans)⇒
Analyze the N-glycan structures by MS analysis, etc. |
Comment 0
|

|
| 8) |
Eluate (de-N-glycosylated peptides)⇒
Identify the respective glycosylation sites in the de-N-glycosylated peptides by subjecting
to automated nanoLC-ESI-MS/MS analysis etc. (by digestion with PNGaseF, Asn in the original glycosylated peptides changed to Asp in de-N-glycosylated peptides (see Fig.1)). |
Comment 0
|
|
|
|
3. |
In gel deglycosylation of N-linked glycoprotein with PNGase F (Kawasaki N et al, 2009). |
| 1) |
Apply a glycoprotein sample (or a sample containing (a) glycoproteins(s) (> 2–10 μg target protein) onto SDS-PAGE. |
Comment 0
|

|
| 2) |
Stain the protein bands with colloidal Coomassie brilliant blue. |
Comment 0
|

|
| 3) |
Excise the target band from the gel. |
Comment 0
|

|
| 4) |
Reduce the excised gel band with 100 μL of 50 mM dithioerythreitol. |
Comment 0
|

|
| 5) |
Alkylate the excised gel band with 65 mM iodoacetamide in 100 μL of 100 mM ammonium bicarbonate buffer, pH 8.5. |
Comment 0
|

|
| 6) |
Destain the gel with 50% acetonitrile in 100 μL of 50 mM ammonium bicarbonate buffer, pH 8.5, two times. |
Comment 0
|

|
| 7) |
Incubate the gel at 37°C overnight with PNGase F (1 mU) in 30 μL of 50 mM ammonium bicarbonate buffer, pH 8.5. |
Comment 0
|

|
| 8) |
Extract the gel additionally several times with water. |
Comment 0
|

|
| 9) |
Combine the released N-linked glycans from the gel. |
Comment 0
|

|
|
|
|
4. |
|
| 1) |
Dissolve 15 mg of lgG (corresponding to 200 nmol of oligosaccharide moiety) in 60 μL of distilled water. |
Comment 1
|

|
| 2) |
Heat at 100°C for 10 min to denature the protein. |
Comment 0
|

|
| 3) |
Adjust the pH of the reaction mixture to 4. |
Comment 0
|

|
| 4) |
Add pepsin (150 μg) and 2 mU of glycopeptidase A. |
Comment 1
|

|
| 5) |
Incubate at 37°C overnight for proteolysis and releasing of oligosaccharide moiety. |
Comment 0
|
|
|
| Notes | Two different N-glycanases, PNGase F and glycopeptidase A are now commonly used as a tool for liberating native oligosaccharides from the original glycoprotein molecule.
The specificity of PNGase F is well established and the enzyme hydrolyzes asparagine-linked glycans representing all major oligosaccharides classes, but both the amino and carboxyl groups of the asparagine residue have to be in peptide linkage. PNGase F preferred a tripeptide or longer (Fan JQ, Lee YC. 1997). Asparagine-linked oligosaccharides containing an α(1→6)-fucose substituted on the asparagine-proximal GlcNAc residue are easily hydrolyzed by PNGase F, but the corresponding core α(1→3)-fucose substituent, found in glycoproteins from plants, insects and other lower animals completely blocks deglycosylation. This appears to be the only known structural feature of an oligosaccharide moiety that confers resistance to PNGase F. In contrast, glycopeptidase A can hydrolyze asparagine-linked oligosaccharides containing an α(1→3)-, as well as α(1→6)-fucose on the asparagine-proximal GlcNAc residue. However, for glycopeptidase A to work well, the size of the peptides must be in the range of 3–40 amino acids and protease digestion of the glycoprotein is required (see Comment in Step 4 of Procedure 4. Thus PNGase F is the most versatile and widely used tool to de-N-glycosylate glycoproteins and/or glycopeptides for further analyses, provided the glycoprotein sources are not from lower animals or plants known or suspected to carry core α(1→3)-fucosylation. In the latter cases, the use of Glycopeptidase A, either directly on glycopeptides or following a prior digestion with PNGase F, is required to obtain both pools of N-glycans with and without core α(1→3)-fucosylation.
PNGases are used as tools for
① investigating whether a glycoprotein has N-glycans or not.
② investigating structure -function studies of biologically active glycoproteins.
③ analyzing the structures of the glycans liberated from the original glycoprotein.
Standard conditions for case ① ; use 1mU(IU) PNGaseF for 25 μg of the denatured glycoprotein (see comment 1), and incubate at 37°C overnight; for case ②; use 10 mU(IU) PNGaseF for 25 μg of the undenatured glycoprotein and incubate at 37°C overnight. Susceptibility of the glycoproteins to PNGaseF are different depending on the glycoprotein sample, check the deglycosylation of an aliquots of the sample, and add the enzyme and incubate for one more night; for case③; use 1mU(IU) PNGaseF for 25 μg of the denatured glycoprotein overnight. In case the detergents added for the denaturation may disturb further structural analysis, deglycosylate the sample in non-denatured condition. |
| Figure & Legends |
Figure & Legends 
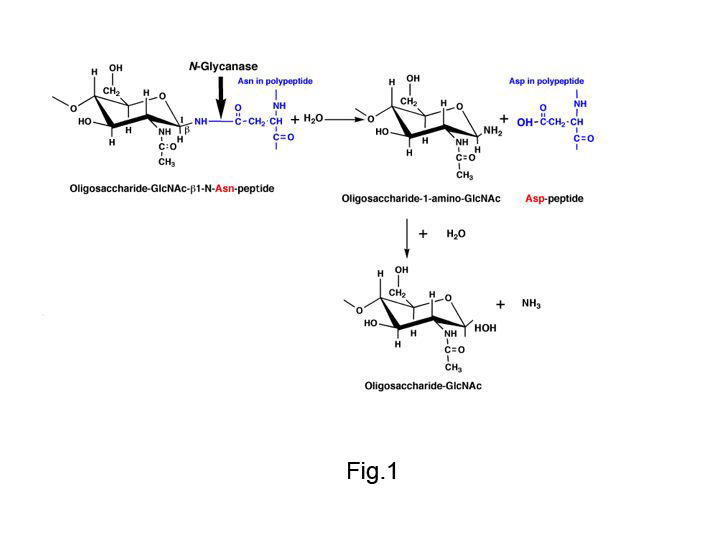
Fig. 1. Reaction mechanisms of N-glycanase (PNGase F and glycopeptidase A)
N-glycanase hydrolyzes the amide bond of β-aspartylglycosylamine and produces glycosylamine, which is subsequently hydrolyzed non-enzymatically.

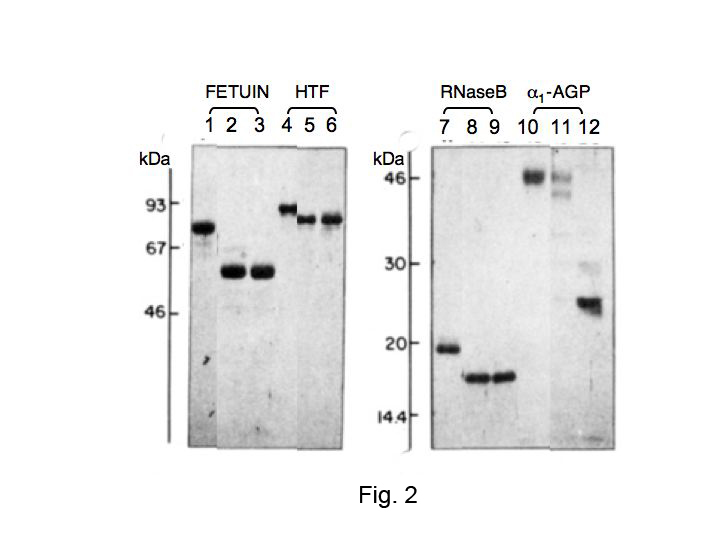
Fig. 2. Comparison of the deglycosylation efficiency of PNGaseF on some N-linked glycoproteins
Glycoprotein stocks (5 mg/mL) were made in either 0.05 M NaCl (native) or 1% SDS and boiled for 3 min (denatured). PNGase F reactions were conducted in 0.25 M sodium phosphate, pH 8.6, with 6 mU of enzyme. Reactions of 100 mL contained 50 μg of test protein and the following: (1) for native, 10 mM β-mercaptoethanol; (2) for denatured, 0.6% NP-40. Incubations were conducted at 37°C for 18 h. Controls (no enzyme), lanes 1, 4, 7, and 10. PNGase F on the native glycoproteins, lanes 2, 5, 8, and 11. PNGase F on the denatured glycoproteins, lanes 3, 6, 9, and 12. Fetuin and human transferrin (HTF) were resolved on 10% gel; ribonuclease B (RNase B) and α1-acid glycoprotein (α1-AGP), resolved on 12.5% gel. Approximately 2 μg proteins per well were applied. [Note: These conditions described in this legend were chosen to demonstrate complete deglycosylation of all susceptible asparagine-linked oligosaccharides. However, depending on the glycoprotein and whether it is in its native conformation or has been unfolded by denaturation, the actual amount of PNGase F required may be much lower (0.6–30 mU/mL). ] Cited from Tarentino AL et al. 1985 with modifications.
This figure was originally published in "Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F" Tarentino AL. et al. [Biochemistry. 24(17):4665–71.] 1985 Aug. DOI:10.1021/bi00338a028.

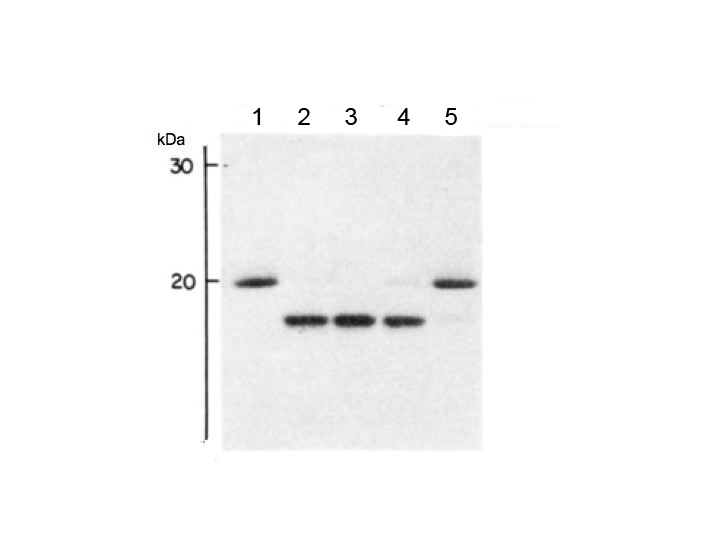
Fig. 3. Deglycosylation of denatured RNase B as a function of added PNGase F
Serial dilutions of a stock PNGaseF (1200 mU/mL) were prepared in 0.1 M sodium phosphate, pH 8.6, and added to 50 μg denatured RNaseB as indicated: lane 1; control (no enzyme), lane 2; 120 mU/mL, lane 3; 12 mU/mL, lane 4; 0.6 mU/mL, lane 5; 0.06mU/mL. The incubation time was 1 h. Conditions for SDS-PAGE as in Fig. 2. Cited from Tarentino AL et al. 1985 with modifications.
This figure was originally published in "Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F" Tarentino AL. et al. [Biochemistry. 24(17):4665–71.] 1985 Aug. DOI:10.1021/bi00338a028.
|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2016-02-10 13:24:57 |
- Fan, J.Q., and Lee, Y.C. (1997) Detailed studies on substrate structure requirements of glycoamidases A and F. J Biol Chem. 272, 27058–27064 [PMID : 9341145]
- Kawasaki, N., Lin, C.W., Inoue, R., Khoo, K.H., Kawasaki, N., Ma, B.Y., Oka, S., Ishiguro, M., Sawada, T., Ishida, H., Hashimoto, T., and Kawasaki, T. (2009) Highly fucosylated N-glycan ligands for mannan-binding protein expressed specifically on CD26 (DPPVI) isolated from a human colorectal carcinoma cell line, SW1116. Glycobiology 19, 437–450 [PMID : 19129245]
- Makino, M., Kojima, T., and Yamashina, I. (1966) Enzymatic cleavage of glycopeptides. Biochem Biophys Res Commun 24, 961–966 [PMID : 5970530]
- Plummer, T.H. Jr., Elder, J.H., Alexander, S., Phelan, A.W., and Tarentino, A.L. (1984) Demonstration of peptide:N-glycosidase F activity in endo-β-N-acetyl- glucosaminidase F preparations. J Biol Chem. 259, 10700–10704 [PMID : 6206060]
- Takahashi, N., and Nishibe, H. (1978) Some characteristics of a new glycopeptidase acting on aspartylglycosylamine linkage. J Biochem. 84, 1467–1473 [PMID : 738997]
- Takahashi, N., Yagi, K., and Kato, K. (2008) Release of N-glycans by enzymatic methods: In Experimental Glycoscience: Glycochemistry, Taniguchi N. et al (eds): pp.7–11, Springer Japan [PMID : not available]
- Tarentino, A.L., and Plummer, T.H. Jr. (1994) Enzymatic deglycosylation of asparagine- linked glycans; purification, properties, and specificity of oligosaccharide-cleaving enzymes from Flavobacterium meningosepticum. Methods Enzymol. 230, 44–57 [PMID : 8139511]
- Tarentino, A.L., Gomez, C.M., and Plummer, T.H. Jr. (1985) Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry 24, 4665–4671 [PMID : 4063349]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Kawasaki, Nobuko,
(2016). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.24,4,2024 .
How to Cite this Work in Website:
Kawasaki, Nobuko,
(2016).
N-Glycanase digestion.
Retrieved 24,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t58.
html source
Kawasaki, Nobuko,
(2016).
<b><em>N</em>-Glycanase digestion</b>.
Retrieved 4 24,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t58" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t58</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|