Sialidase enzymes catalyze the removal of sialic acid residues from glycoproteins, glycolipids and oligosaccharides. Four types of mammalian sialidases (Neu1, Neu2, Neu3, Neu4) have been characterized in the terms of their subcellular localization and enzymatic properties, all being involved in multiple cellular functions. Activity assays therefore provide important information for understanding how sialidases actually influence various biological events. |
| Category | Glycosidases & related proteins |
| Protocol Name | Enzyme assay of sialidases |
Authors
 |
Miyagi, Taeko
Division of Cancer Glycosylation Research,Institute of Molecular Biomembrane and Glycobiology, Tohoku Pharmaceutical University
|
| KeyWords |
|
Reagents
 |
| ● |
Buffers : Sodium acetate buffer (pH 4.5 or 5.5), Sodium phosphate buffer (pH 6.5) |
| ● |
Substrates : 4-methylumbelliferyl N-acetylneuraminic acid (4MU-NeuAc, Nacalai Tesque, Inc., Kyoto, Japan) |
| ● |
Substrates : Gangliosides, Mixed gangliosides from bovine brain or GM3 (NeuAcα2-3Galβ1-4Glcβ1-1Cer) (Calbiochem, San Diego, CA) |
| ● |
Triton X-100 (Calbiochem) |
| ● |
Bovine serum albumin (Fraction V, Sigma-Aldrich, St. Louis, MO) |
| ● |
|
| ● |
4-methylumbelliferylone (4MU) (Nacalai Tesque, Inc.) |
| ● |
|
| ● |
|
| ● |
TBA (thiobarbituric acid) |
| ● |
|
| ● |
|
| ● |
|
| ● |
|
| ● |
|
| ● |
Ammonium acetate buffer (pH 5.5) or 1,2-Diamino-4, 5-methylenedioxybenzene (DMB) kit (Takara Bio Inc., Otsu, Japan) |
|
Instruments
 |
| ● |
|
| ● |
|
| ● |
|
| ● |
|
| ● |
|
| ● |
Reverse phase Luna 5u C18 column (Phenomenex, Torrance, CA) |
|
| Methods |
|
1. |
Sialidase activity assays with ganglioside substrates |
| 1) |
Place the reaction mixture in a test tube.
Gangliosides (releasable sialic acid sites) 5 nmol
TritonX-100 or sodium cholate 50 μg
Bovine serum albumin 50 μg
Enzyme ( to 0.5 mg protein)
in 50 μL |
Comment 1
|

|
| 3) |
Determine released sialic acids by TBA or HPLC methods. |
Comment 0
|
|
|
|
2. |
Determination of released sialic acids (TBA method (Warren 1963)) |
| 1) |
Terminate the reaction (50 μL) by adding 25 μL metaperiodate solution. |
Comment 1
|

|
| 2) |
Stand for 20 min at room temperature. |
Comment 0
|

|
| 3) |
Add 250 μL arsenite solution and vortex the tube until the yellow-brown colour
disappears. |
Comment 1
|

|
| 4) |
Add 750 μL TBA solution, vortex and heat in a boiling water bath for 15 min. |
Comment 1
|

|
| 6) |
Add 1 mL cyclohexanone, vortex and centrifuge at 3,000 rpm for 10 min. |
Comment 0
|

|
| 7) |
Measure the upper phase at 549 nm. |
Comment 0
|
|
|
|
3. |
HPLC analysis with fluorimetric detection ( Li, 1992 ) |
| 1) |
Add 50 μL malononitrile (0.8%) to the reaction mixture (25 μL). |
Comment 1
|

|
| 2) |
Add 355 μL tetraborate (0.15 M, pH 9.5). |
Comment 0
|

|
| 4) |
Apply 10–20 μL to a reversed-phase column (C18) and separate with a methanol and
ammonium acetate buffer (15:85 v/v, pH 5.5). |
Comment 0
|

|
| 5) |
Measure fluorometrically. (emission 434 nm, excitation 357 nm) |
Comment 0
|
|
|
|
4. |
Sialidase activity assays with the 4MU-NeuAc substrate. |
| 1) |
Pprepare the reaction mixture.
Sodium acetate buffer (pH 4.5 or pH 5.5) 10 μmol
4MU-NeuAc 20 nmol
Bovine serum albumin 0.1 mg
Enzyme
in 0.1 mL |
Comment 0
|

|
| 3) |
Terminate the reaction by addition of 1.25 mL 0.25M glycine-NaOH (pH 10.4). |
Comment 0
|

|
| 4) |
Measure fluorometrically the 4-methylumbelliferylone(4-MU) released. (emission 448 nm, excitation 365 nm) |
Comment 0
|
|
|
| Notes | Four types of mammalian sialidases have been identified and characterized to date, designated as Neu1, Neu2, Neu3 and Neu4. They differ in enzymatic properties and subcellular sites as well as in their primary structures and chromosomal localization (Miyagi, et al. 2008). All contain several Asp boxes (-Ser-X-Asp-X-Gly-X- Thr-Trp-) and the Arg-Ileu-Pro sequence, which are conserved sequences also found in sialidases from microorganisms, despite the fact that the mammalian enzymes do not exhibit any other sequence similarities to microbial sialidases. Among human sialidases, the overall amino acid identity of NEU1 to the other forms is relatively low (19–24%), while NEU2, NEU3 and NEU4 show 34–40% homology to each other. Regarding comparative expression levels of human sialidases, NEU1 generally shows the highest expression, 10–20 times higher than those of NEU3 and NEU4, while NEU2 expression is extremely low, only one four- to ten- thousandth of the NEU1 value at the most in a wide range of tissues, as assessed by quantitative real time RT-PCR. However, these profiles differ among the human, rat and mouse. With regard to subcellular localization, sialidases can functionally move to sites other than their major locations, such as to the cell surface, depending on physiological conditions.
Neu1 mainly localized in lysosomes is a target gene for sialidosis (sialidase deficiency), and is associated with a protective protein (carboxypeptidase A) and β-galactosidase as a complex in lysosomes, dissociation of the complex leading to sialidase inactivation. With an optimum pH of about 4.5, Neu1 desialylates mainly glycopeptides and oligosaccharides, as well as 4MU-NeuAc, a synthetic substrate. The cytosolic sialidase, Neu2, shows an optimum pH of about 5.5–6.5 and can desialylate fetuin and gangliosides as well as 4MU-NeuAc. The plasma membrane-associated sialidase, Neu3, is essentially specific for gangliosides other than GM1 and GM2, in the presence of Triton X-100. NEU3 is involved in regulation of cellular signaling, and its expression is markedly increased in various human cancers, leading to enhancement of malignant properties including suppression of apoptosis. Neu4 has an acidic optimal pH at about pH 4.5. The human ortholog, NEU4, has been reported to be localized in two different sites: one is the lysosomal lumen, and the other being the mitochondria and microsomal membranes. NEU4 appears to consist of iso-forms differing in the presence and absence of 12 N-terminal amino acid residues that act to target mitochondria, so that this might explain variation in localization. The isoforms are also differentially expressed in a tissue-specific manner, brain, muscle and kidney containing both, and the liver and colon possessing predominantly the short form. Unlike other human sialidases, the enzyme possesses broad substrate specificity, with sensitivity to mucin. |
| Figure & Legends |
Figure & Legends 
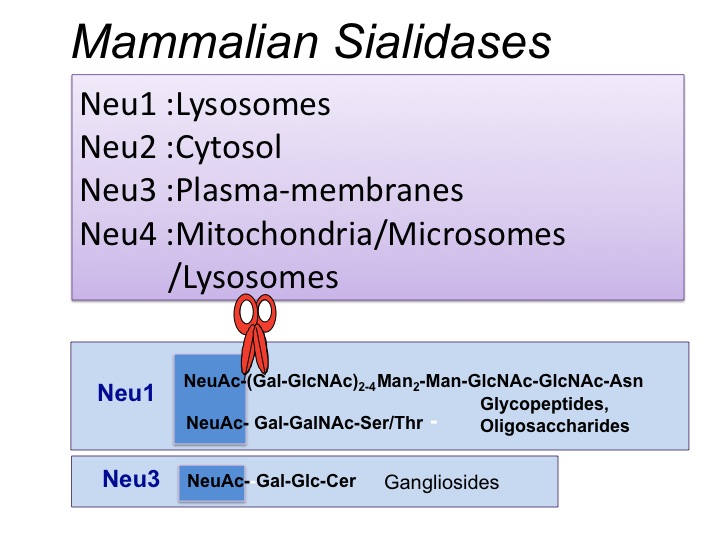
Fig. 1. Four types of mammalian sialidases. They have been designated as Neu1, Neu2, Neu3, and Neu4, and differ in subcellular localization and enzymatic properties
Neu1 preferentially hydrolyzes oligosaccharides and glycopeptides but hardly gangliosides. Neu3 is a key enzyme for ganglioside degradation because of its strict substrate preference for gangliosides, which co-localize with this sialidase in surface membranes. Neu1 and Neu3 possess an acidic optimal pH for their activity. Neu2 and Neu4 can act on gangliosides as well as oligosaccharides and glycoproteins at nearly neutral pH and acidic pH, respectively.

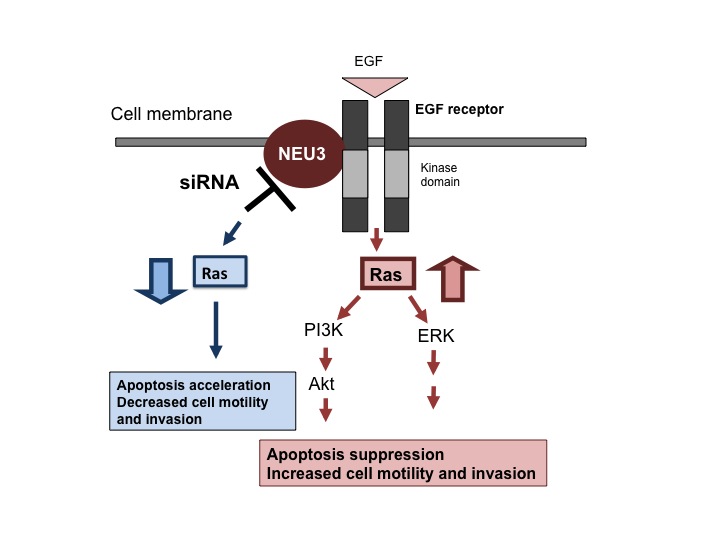
Fig. 2. Roles of the human Neu3 ortholog, NEU3, in apoptosis of cancer cells via the epidermal growth factor receptor
NEU3 siRNA inhibits and NEU3 overexpression stimulates Ras activation, with consequent influence on ERK and Akt through modulation of tyrosine-phosphorylation of the epidermal growth factor receptor.
This figure was originally published in J Biochem. 144(3):279–85. 2008 "Plasma membrane-associated sialidase as a crucial regulator of transmembrane signalling" Miyagi T. et al. Oxford University Press.
|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-01-06 15:51:57 |
- Hara, S., Yamaguchi, M., Takemori, Y., Nakamura, M. and Ohkura, Y. (1986) Fluorometric high-performance liquid chromatography of N-acetyl- and N-glycolylneuraminic acids and its application to their microdetermination in human and animal sera, glycoproteins, and glycolipids. J. Chromatogr. 377, 111–119 [PMID : 3711202]
- Li, K. (1992) Determination of sialic acids in human serum by reversed-phase liquid chromatography with fluorimetric detection. J. Chromatogr. 579, 209–213 [PMID : 1429968]
- Miyagi, T. and Tsuiki, S. (1985) Purification and characterization of cytosolic sialidase from rat liver. J. Biol. Chem. 260, 6710–6716 [PMID : 3997847]
- Miyagi, T., Wada, T., Yamaguchi, K., Hata, H., and Shiozaki, K. (2008) Minireview; Plasma membane-associated sialidase as a crucial regulator of transmenbrane signaling. J. Biochem. 144,279–285 [PMID : 18632803]
- Warren, L. (1963) Thiobarbituric acid assay of sialic acids "Methods in Enzymology", ed. by Colowick S. P., Kaplan, N.O., Academic Press, New York, Vol. 6 p.463 [PMID : not found]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Miyagi, Taeko,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.25,4,2024 .
How to Cite this Work in Website:
Miyagi, Taeko,
(2015).
Enzyme assay of sialidases.
Retrieved 25,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t42.
html source
Miyagi, Taeko,
(2015).
<b>Enzyme assay of sialidases</b>.
Retrieved 4 25,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t42" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t42</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|