Sialyltransferases (EC No. 2.4.99) catalyze the transfer of sialic acid from the nucleotide sugar donor CMP-Sia to acceptor oligosaccharides found on glycoproteins, polysaccharides, and glycolipids. In this section, the enzymes transfer siallic acid to glycoproteins and polysaccharides are mainly mentioned. The details about the enzymes of ganglioside synthesis, see (Enzyme assay of glycolipide glycosyltransferases). |
| Category | Glycosyltransferases & related proteins |
| Protocol Name | Enzyme assay of sialyltransferases for oligosaccharides |
Authors
 |
Tsuji, Shuichi
Institute of Glycoscience, Tokai University
|
| KeyWords |
|
Reagents
 |
| ● |
CMP-[6-14C]NeuAc or CMP-[6-3H]NeuAc (GE Healthcare, Little Chalfont, UK) |
| ● |
|
| ● |
NANase I (specific for a2,3-linked sialic acids units/mL: Glyko/Prozyme, Inc., Hayward, CA) (if necessary) |
| ● |
NANase II (specific for a2,3-and a2,6-linked sialic acids 5 units/mL: Glyko/Prozyme, Inc.) (if necessary) |
| ● |
V. cholerae sialidase (specific for a2,3-, a2,6-linked and a2,8-linked sialic acids 1 unit/mL: Roche Diagnostics, Basel, Switzerland) (if necessary) |
| ● |
Newcastle disease virus sialidase (specific for a2,3- and a2,8-linked sialic acids 5 units/mL: Oxford Glycosystems Ltd., Oxon, UK) (if necessary) |
|
Instruments
 |
| ● |
Fuji BAS2000 radio image analyzer |
|
| Methods |
|
1. |
|
| 1) |
Enzyme activity is measured in 50 mM MES buffer (pH 6.0), 1 mM MgCl2, 0.5% Triton CF-54, 100 mM CMP-[14C]NeuAc, 10 μg of oligosaccharides, and enzyme preparation, in a total volume of 10 μL. |
Comment 0
|
|
|
|
2. |
|
| 1) |
The enzyme reaction is performed at 37°C for 3–20 h. |
Comment 0
|
|
|
|
3. |
Reaction termination and product detection |
| 1) |
directly subjected to HPTLC with a solvent system (for example: 1-propanol, aqueous ammonia, and water (6:1:2.5)). |
Comment 0
|

|
| 2) |
The radioactive materials are visualized and quantified with a Fuji BAS2000 radio image analyzer. |
Comment 1
|
|
|
|
4. |
Linkage analysis of incorpotared sialic acids (if necessary) |
| 1) |
digest [14C]NeuAc -incorporated productswith a linkage-specific exosialidase. |
Comment 0
|

|
| 2) |
subject to HPTLC with a solvent system (for example: 1-propanol/aqueous ammonia/water (6:1:2.5)). |
Comment 0
|

|
| 3) |
further procedure is essentually the sama as that of 3-2). |
Comment 0
|
|
|
| Notes | comments for reagent
1) CMP-Sia
If the acceptor substrate is labeled with isotope or some fluorescent materials, you can use cold CMP-Sia. If not, you should use isotope labeled CMP-Sia, such as CMP-[6-14C]NeuAc or CMP-[6-3H]NeuAc from Amasham. In this assay, you should use the highest purified CMP-Sia, such as Cytidine-5'-monophospho-N-acetylneuraminic Acid Disodium Salt (Nacalai Tesque code no: 10432-24).
2) Detergent
The sialyltransferase itself does not require detergent for the enzymatic activity. If the solubility of acceptor substrate is poor, you should use detergent, such as Triton CF-54. Although Triton CF-54 was commonly used as a detergent for the enzyme assay, unfortunately, it is now hardly obtainable. As far as seen our data, Triton CF-74 is well substituted for Triton CF-54.
3) Other reagents used in this assay should be of the highest purity obtainable.
comments for procedure
This assay protocol is mainly for mammalian enzyme. Though it will be essentially adaptable for other enzymes, if you try to assay other enzymes, please check the following references.
1) Mammalian sialyltransferases (Takashima et al, 2002)
From the vertebrate, 20 sialyltransferase genes and their essential genomic structure have been identified, respectively. Their cDNAs have been cloned and their enzymatic properties have been characterized (Table 1).
The cloned sialyltransferases have a type II transmembrane topology and contain highly conserved domains called sialylmotifs L (Long), S (Short), VS (Very Short) and III. These motifs are highly conserved regions comprising about 20% of the total protein sequence. The L- and S-sialylmotifs were shown to bind to the donor CMP-NeuAc and acceptor saccharide substrates. The VS-sialylmotif is considered to be participated in the catalytic center though the strict data has not been published. Each sialyltransferase exhibits strict specificity for acceptor substrates and linkages they synthesize.
The cloned sialyltransferases can be classified into four families according to the carbohydrate linkages they synthesize, i.e., the b-galactoside α2,3-sialyltransferase family (ST3Gal-I-VI), the β-galactoside α2,6-sialyltransferase family (ST6Gal-I,II), the GalNAc α2,6-sialyltransferase family (ST6GalNAc-I-VI), and the α2,8-sialyltransferase family (ST8Sia-I-VI).
2) Bacterial sialyltransferases
Several bacterial sialyltransferases (ST3Gal, ST6Gal and ST8Sia) have been cloned and characterized (Yamamoto et al., 1998). Interestingly, the sialylmotif has not been found in the amino acid sequences of these bacterial sialyltransferases even though they transfer NeuAc from CMP-NeuAc to oligosaccharides in analogous to the mammalian enzymes. Generally, bacterial enzymes have broader acceptor tolerances compared to mammalian enzymes. For example, Neisseria meningitidis ST3Gal can utilize both α- and (3-linked terminal galactoses as acceptors (Gilbert et al., 1997).
3) Viral sialyltransferases
A novel myxoma virus early gene, MST3N, is a member of the eukaryotic sialyltransferase gene family. Myxoma virus is a member of the poxvirus family of double stranded DNA viruses, and it is known to infect Old World or European rabbits causing highly lethal myxomatosis. mammalian cells infected by Myxoma virus produce viral α2,3-sialyltransferase (v-ST3Gal-I) in the cells. v-ST3Gal-I is closely related to mammalian ST3Gal-IV, though it has a very broad acceptor specificity that is not found among the mammalian or bacterial α2,3-sialyltransferases. Acceptors include not only type I to III disaccharides but also fucosylated Lewisx and Lewisa (Sujino, K. et al., 2000)
4) Plant sialyltransferases (Takashima et al, 2006)
Sialic acids are widely distributed among living creatures, from bacteria to mammals, but it has been commonly accepted that they do not exist in plants, although the possibility remains that sialylated glycoconjugates may exist in plant cells. Interestingly, however, putative gene homologs for mammalian sialyltransferases and CMP-Sia transporters have been detected in the genome and/or expressed sequence tag (EST) databases of some plants, such as Arabidopsis thaliana (thale-cress) and Oryza sativa (Japanese rice). This suggests that plants have potential ability of sialylation despite the absence of sialic acids in them. Takashima et al. cloned three genes from 0. sativa, each encoding a protein having sialyl motif-like sequences, and analyzed the enzymatic activity of the proteins. One of them, called OsSTLP1, really transferred sialic acid from CMP-NeuAc to galactose of Galβ1,4GlcNAc through α2,6-linkage. |
| Figure & Legends |
Figure & Legends

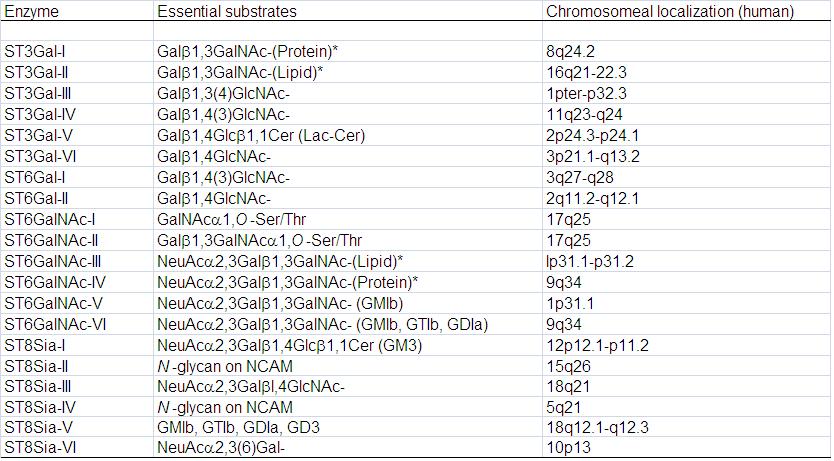
Table 1. Acceptor specificities of mouse and human sialyltransferases, and chromosomal localization of human genes
Asterisk means preferential but not specific substratre.
This table was originally published in J Biol Chem. Takashima S, Tsuji S. et al. "Characterization of the second type of human beta-galactoside alpha 2,6-sialyltransferase (ST6Gal II), which sialylates Galbeta 1,4GlcNAc structures on oligosaccharides preferentially. Genomic analysis of human sialyltransferase genes" 2002, 277(48):45719-28, and reused in "Experimental Glycoscience -Glycobiology" edited by Taniguchi N. et al. Springer Japan KK. 2008, pp.42-46 (Tsuji S. Part1: Section I).
|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-11-09 16:14:28 |
- Sujino, K., Jackson, R.J., W.C.Chan, N., Tsuji, S. and Palcic, M.M. (2000) A novel viral α2,3-sialyltransferase (v-ST3Gal I): transfer ofsialic acid to fucosylated acceptors. Glycobiology 10, 313–320 [PMID : 107045430]
- Takashima, S., Abe, T., Yoshida, S., Kawahigashi, H., Saito, T., Tsuji, S., and Tsujimoto, M. (2006) Analysis of sialyltransferase-like proteins from Oryza sativa. J. Biochem 139, 279–287 [PMID : 16452316]
- Yamamoto,T., Nakashizuka, M., and Terada, I. (1998) Cloning and expression of a marine bacterial b-galactoside α2,6-sialyltransferase gene from Photobacterium damsela JT0160. J. Biochem. 123, 94–100 [PMID : 9504414]
- Gilbert, M., Cunningham, A.-M., Watson, D.C., Martin, A., Richards J.C., and Wakarchuk, W.W. (1997) Characterization of a recombinant Neisseria meningitidis α2,3-sialyltransferase and its acceptor specificity. Eur. J. Biochem. 249, 187–194 [PMID : 9363771]
- Takashima, S., Tsuji, S., and Tsujimoto, M. (2002) Characterization of the second type of human β-galactoside α2,6-sialyltransferase (ST6Gal II) that sialylates Galbl,4GlcNAc structures on oligosaccharides preferentially. J. Biol. Chem. 277, 45719–45728 [PMID : 12235148]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Tsuji, Shuichi,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.23,4,2024 .
How to Cite this Work in Website:
Tsuji, Shuichi,
(2015).
Enzyme assay of sialyltransferases for oligosaccharides.
Retrieved 23,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t37.
html source
Tsuji, Shuichi,
(2015).
<b>Enzyme assay of sialyltransferases for oligosaccharides</b>.
Retrieved 4 23,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t37" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t37</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|