Tunicamycin targets GlcNAc transferase, and prevents assembly of GlcNAc-PP-dolichol and thus assembly of G3M9GlcNAc2-PP-dolichol. The presence of tunicamycin results in proteins missing some or all of their N-linked side chains. Tunicamycin may also inhibit protein synthesis, not all proteins will be affected to the same degree. |
| Category | Biosynthesis & Metabolism |
| Protocol Name | |
Authors
 |
Yong Ma, Bruce
Research Center for Glycobiotechnology, Ritsumeikan University
|
| KeyWords |
|
Reagents
 |
| ● |
Cultured cell line, either adherent or suspension |
| ● |
Complete culture medium appropriate for cell line |
| ● |
Tunicamycin (stock solution 1mg/mL), an inhibitor of N-linked glycosylation: Dissolve tunicamycin (mw 840) in dimethylsulfoxide (DMSO), dimethylformamide (DMF), 95% ethanol, or 25 mM NaOH to 1 mg/mL. Stable ~1 year at −20°C. |
| ● |
Solvent used for making inhibitor solution |
| ● |
Multiply deficient medium without glucose |
| ● |
[3H]mannose (5 to 20 Ci/mmol) |
| ● |
[35S]methionine (> 800 Ci/mmol) |
| ● |
Metabolic radiolabeling of glycoconjugates |
| ● |
Phosphate-buffered saline (PBS) |
| ● |
|
| ● |
0.5 U/mL endoglycosidase H (endo H) and endo H digestion buffer
[Note] All chemicals for TCA precipitation, gel electrophoresis, immunoprecipitation and immunoblotting. |
|
Instruments
 |
| ● |
37°C, 5% CO2 incubator for cell culture |
| ● |
Laser confocal microscopy (e.g. FV1000, Olympus, Tokyo, Japan) |
| ● |
Image analyzer for chemiluminescence (e.g. LAS-4000, Fujifilm, Tokyo, Japan) |
| ● |
|
| ● |
24-well and 100-mm tissue culture plates |
| ● |
1.5-mL, 15-mL, or 50-mL conical polypropylene centrifuge tube |
| ● |
Additional regents and equipment for counting viable cells |
| ● |
|
|
| Methods |
|
1. |
Determine optimal tunicamycin concentration |
| 1) |
For tunicamycin tested, set up 15 wells of a 24-well tissue culture plate with 1.8 mL cells/well (1–5 × 104 cells/well). Incubate 24 h. |
Comment 1
|

|
| 2) |
While cells are incubating, make a series of 1:1 (v:v) dilutions of tunicamycin as follows: Place 80 μL of tunicamycin stock solution (1 mg/mL) in a sterile microcentrifuge tube and dilute to 800 μL with complete culture medium. Transfer 400 μL to a second tube and dilute with 400 μL complete culture medium. Repeat for a total of seven tubes (tunicamycin concentration range 0.15–10 μg/mL). As a control, prepare an identical series of dilutions of the solvent used for making the tunicamycin stock solution. |
Comment 0
|

|
| 3) |
Add 200 μL tunicamycin-containing medium from each dilution tube to a well of the tissue culture plate (from step 2) containing cells (2 mL final). Add 200 μL complete culture medium alone to the remaining well (as a zero-inhibitor control point). |
Comment 1
|

|
| 4) |
Incubate plate 24 h. Store the unused tunicamycin-containing medium (200 μL/tube) at 4°C until used in step 6. |
Comment 0
|

|
| 5) |
Perform short-term labeling of cells with [35S]methionine using 0.2 mCi/mL label and 2 mL final volume per well. |
Comment 0
|

|
| 6) |
Add the remaining 200 μL of each dilution of tunicamycin (from step 4) to the wells and incubate an additional 4 h. |
Comment 0
|

|
| 7) |
Harvest cells and determine the incorporation of [35S]methionine into macromolecules by TCA precipitation. |
Comment 0
|

|
| 8) |
Plot label incorporation versus tunicamycin concentration. |
Comment 1
|
|
|
|
2. |
Label cells in the presence of tunicamycin for inhibiting N-linked glycosylation |
| 1) |
Split a new cell sample into complete medium in a 100-mm tissue culture plate and incubate 24 h. The cell density should be 5 × 105 to 2.5 × 106 cells/plate. |
Comment 0
|

|
| 2) |
Add tunicamycin to the optimal concentration determined above. Set up a control plate containing same quantity of stock solution solvent but no tunicamycin. Incubate 24 h. |
Comment 0
|

|
| 3) |
Wash cells (using the same procedure as employed during short-term labeling, step 1–5) and, to each plate, add sufficient glucose-free MDM to cover the cells (~5 mL for a 100-mm plate). |
Comment 0
|

|
| 4) |
Add tunicamycin or solvent as in step 2-2. Add [3H]mannose to 0.02 to 0.1 mCi/mL. Incubate 4 to 12 h. |
Comment 1
|

|
| 5) |
Harvest adherent cells by scraping with a disposable scraper into ice-cold PBS. Harvest suspension cells by centrifuging several minutes at 300 × g, 4°C, and resuspend in ice-cold PBS. |
Comment 0
|
|
|
|
3. |
Measure incorporated macromolecular radioactivity |
| 1) |
Determine the amount of incorporated macromolecular radioactivity by TCA precipitation. |
Comment 0
|

|
| 2) |
If desired, and if methods are available for purifying and identifying a specific glycoprotein from cells rediolabeled with [35S]methionine, examine the effect of a given tunicamycin by SDS-PAGE and autoradiography. |
Comment 0
|

|
| 3) |
Instead of autoradiography, immunoprecipitation together with immunoblotting can be used for analyzing the inhibition of N-linked glycosylation by tunicamycin. |
Comment 0
|

|
| 4) |
Instead of autoradiography, confocal microscopy can be used for studying the role of tunicamycin on the subcellular localization of cargo transport lectins in glycoprotein quality control. |
Comment 0
|
|
|
| Discussion | The optimal concentration of tunicamycin for the experiment (i.e., highest nontoxic concentration) is determined by monitoring [35S]methionine incorporation as a measure of protein biosynthesis.
The tunicamycin’s ability to inhibit oligosaccharide processing is then determined by analyzing cells labeled with [3H]mannose using TCA precipitation or endoglycosidase H digestion.
Tunicamycin also blocks the assembly of type II keratan sulfate chains, because these glycans also utilize GlcNAc-PP-dolichol. Glycolipid biosynthesis may also be inhibited, although the mechanism underlying this has not been established.
Each N-linked carbohydrate chain contributes ~2000 to 4000 Da to a protein’s mass, so synthesis of a protein in the presence of tunicamycin will result in faster mobility on SDS-PAGE and generally show less size heterogeneity, and proteins will not shift to a faster mobility on SDS-PAGE after PNGase F treatment. |
| Figure & Legends |
Figure & Legends 
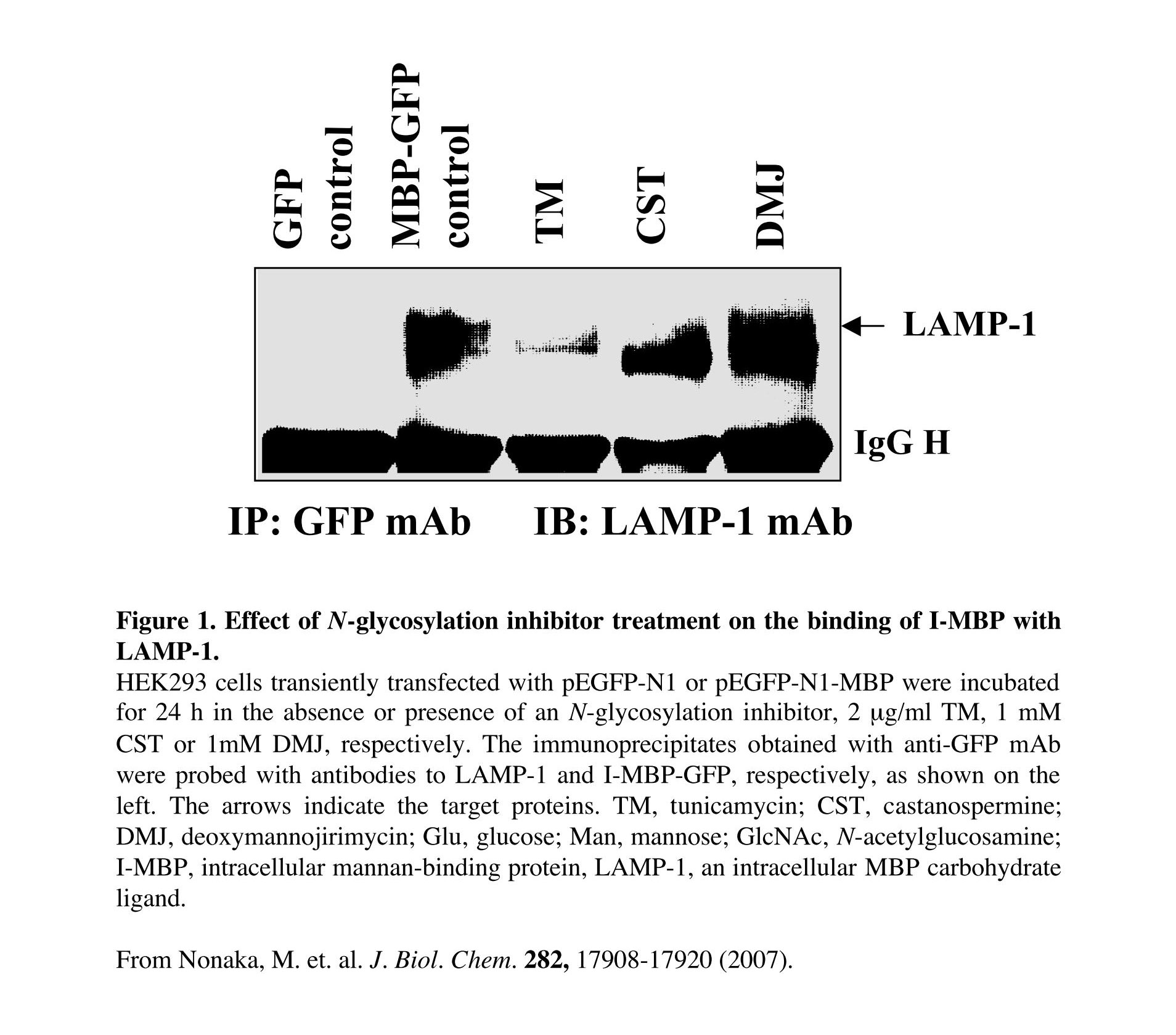
Fig. 1. Effects of N-glycosylation inhibitor treatment on the I-MBP/LAMP-1 interaction
HEK293 cells were treated with the inhibitors for 24 h at 37°C prior to transfection. After transfection, the cells were cultured in the presence of inhibitors again for 24 h under the same conditions prior to harvesting for immunoprecipitation and immunoblotting.
This figure was originally published in J Biol Chem. Nonaka M, Ma BY et al. "Subcellular localization and physiological significance of intracellular mannan-binding protein" 2007 282(24):17908–20. © the American Society for Biochemistry and Molecular Biology.

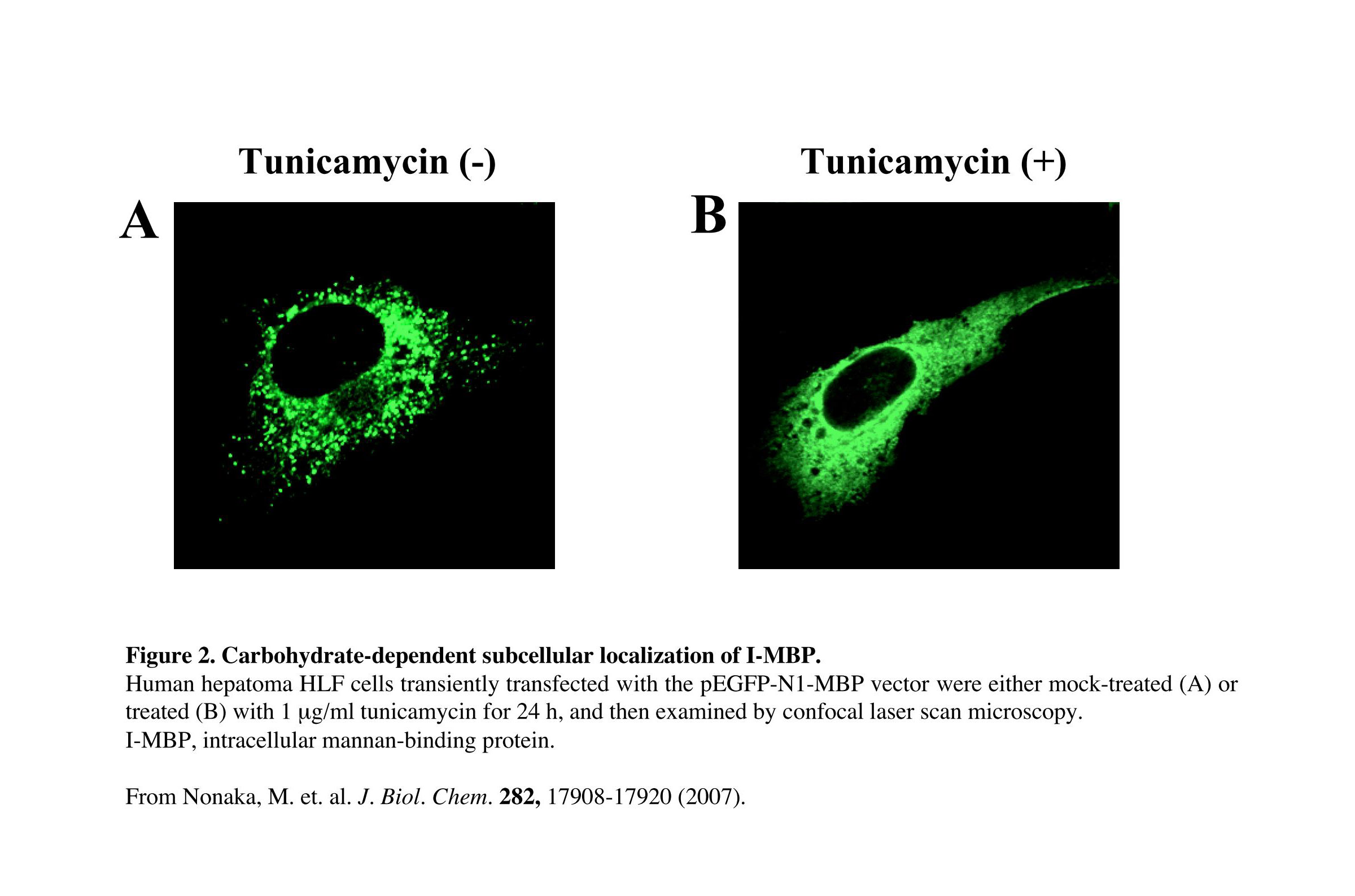
Fig. 2. Carbohydrate-dependent subcellular localization of I-MBP
MBP-GFP-transfected human hepatoma HLF cells were treated for 24 h at 37°C with 1 μg/mL tunicamycin, which prevents N-linked glycosylation. Confocal microscopy showed that the subcellular localization of I-MBP exhibited an equally diffuse cytoplasmic distribution (Fig. 2B), in contrast to the punctate structures observed in the control cells (Fig. 2A).
This figure was originally published in J Biol Chem. Nonaka M, Ma BY et al. "Subcellular localization and physiological significance of intracellular mannan-binding protein" 2007 282(24):17908–20. © the American Society for Biochemistry and Molecular Biology. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-02-03 14:33:49 |
- Nonaka, M., Ma, B.Y., Ohtani, M., Yamamoto, A., Murata, M., Ito, Y., Miwa, K., Nogami, W., Kawasaki, N., and Kawasakim T. (2007) Subcellular localization and physiological significance of intracellular mannan-binding protein. J. Biol. Chem. 282, 17908–17920 [PMID : 17442667]
- Elbein, A.D. (1987) Inhibitors of the biosynthesis and processing of N-linked oligosaccharide chains. Annu. Rev. Biochem. 56, 497–534 [PMID : 3304143]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Yong Ma, Bruce,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.26,4,2024 .
How to Cite this Work in Website:
Yong Ma, Bruce,
(2015).
Tunicamycin.
Retrieved 26,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t30.
html source
Yong Ma, Bruce,
(2015).
<b>Tunicamycin</b>.
Retrieved 4 26,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t30" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t30</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|