O-glycosylation represents a very diverse group of modifications, including the O-GalNAc type typical of mucin and the O-GlcNAc, O-Fuc, O-Man and O-Xyl types. These modifications not only activate proteins, but also serve as markers indicating binding to other proteins. However, characterization of O-glycosylation of proteins has remained challenging because no analogous endoglycosidase is known, and alternative chemical protocols are often either incomplete, lack specificity, or degrade one or both products (i.e., the glycan and/or the peptide/protein). Recently, we developed a novel one-pot O-glycome analytical method combined with release from glycoproteins and labeling with pyrazolone analogs. This method, namely β-elimination in the presence of pyrazolone analogs (BEP), allows simultaneous labeling of released O-glycans with pyrazolone analogs and, thus, minimizes the undesirable peeling reaction. Furthermore, BEP allows analysis of not only released O-glycans but also formerly O-glycosylated glycopeptides, because both species can be labeled by pyrazolone analogs and are thus amenable to further analysis. |
| Category | O-Glycans |
| Protocol Name | O-glycosylation analysis by β-elimination in the presence of pyrazolone analogue (BEP) |
Authors
 |
Furukawa, Jun-ichi
Laboratory of Medical and Functional Glycomics, Graduate School of Advanced Life Science and Frontier Research Center for Post-Genome Science and Technology, Hokkaido University
Shinohara, Yasuro
*
Laboratory of Medical and Functional Glycomics, Graduate School of Advanced Life Science and Frontier Research Center for Post-Genome Science and Technology, Hokkaido University
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
1-phenyl-3-Methyl-5-Pyrazolone (PMP) (Tokyo Chemical Industry Co., Ltd., Tokyo, Japan) |
| ● |
1 mol/L sodium hydroxide solution (Wako Pure Chemical Industries, Ltd., Osaka, Japan) |
| ● |
methanol (Wako Pure Chemical Industries, Ltd.) |
| ● |
1 mol/L hydrochloric acid (Wako Pure Chemical Industries, Ltd.) |
| ● |
chloroform (Wako Pure Chemical Industries, Ltd.) |
| ● |
acetic acid (Wako Pure Chemical Industries, Ltd.) |
| ● |
acetonitrile (Wako Pure Chemical Industries, Ltd.) |
| ● |
trifluoroacetic acid (Wako Pure Chemical Industries, Ltd.) |
| ● |
dihydroxybenzoic acid (Sigma-Aldrich, St. Louis, MO) |
|
Instruments
 |
| ● |
Bio-spin disposable chromatography column (Bio-Rad Laboratories, Hercules, CA) |
| ● |
Ultrasonic Homogenizer (TAITEC Co., Ltd., Koshigaya, Japan) |
| ● |
T 100TM Thermal cycler (Bio-Rad Laboratories) |
| ● |
Iatrobeads 6RS-8060 (LSI Medience Corporation, Tokyo, Japan) |
| ● |
Carbograph 1SPE 120/400 MESH (Grace/Alltech Inc., Brentwood, TN) |
| ● |
centrifugal evaporator CVE-3100 (Tokyo Rikakikai Co., Ltd., Tokyo, Japan) |
| ● |
Ultraflex II (Bruker Daltonics Inc., Billerica, MA) |
|
| Methods |
|
1. |
Release and labeling of O-glycan from glycoprotein by β-elimination in the presence of pyrazolone analogues (BEP). |
| 1) |
10 μL of glycoprotein solution in PCR tube. |
Comment 1
|

|
| 3) |
Add 20 μL of PMP reagent in methanol. (0.5 M) |
Comment 0
|

|
| 5) |
BEP reaction
Heat at 85ºC, 16 h. (see Comment)
For glycosylation site analysis, jump to Step 3). |
Comment 0
|

|
| 6) |
Neutralize the reaction mixture with 1 M hydrochloric acid. |
Comment 0
|

|
| 7) |
Add chloroform and mix vigorously. |
Comment 0
|

|
| 9) |
Repeat Step 7) and 8) three times. |
Comment 0
|
|
|
|
2. |
Purification of bisPMP labeled O-glycans |
| 1) |
Prepare 50 mg of graphitized carbon in a bio-spin column. |
Comment 1
|

|
| 2) |
Wash the column with 0.05% trifluoroacetic acid (TFA) in 80% acetonitrile/ 20% Milli-Q water, Milli-Q water, and equilibrate with 0.05% TFA in 10% acetonitrile/ 90% Milli-Q water. |
Comment 0
|

|
| 3) |
Load the above prepared samples onto column. |
Comment 0
|

|
| 4) |
Wash the column sequentially with 200 μL of Milli-Q water, 200 μL of 0.05% TFA in 40% acetonitrile/ 90% Milli-Q water, and 2 mL of 0.05% TFA in 20% acetonitrile/ 80% Milli-Q water. |
Comment 0
|

|
| 5) |
Elute the bisPMP labeled O-glycans with 600 μL of 0.05 % TFA in 40% acetonitrile/ 60% Milli-Q water. |
Comment 0
|

|
| 6) |
Concentrate the sample using speedvac for 1 h at 40°C and resolve in 90% acetonitrile/ 10% Milli-Q water. |
Comment 0
|

|
| 7) |
Prepare 50 mg of iatrobeads in a bio-spin column. |
Comment 1
|

|
| 8) |
Wash the column with 1 M acetic acid and acetonitrile. |
Comment 0
|

|
| 9) |
Load the prepared samples in the Step 2–6 onto the column. |
Comment 0
|

|
| 10) |
Wash the column sequentially with 200 μL of 1% acetic acid in 95% acetonitrile/ 4% Milli-Q water and 200 μL of 2% acetic acid in acetonitrile. |
Comment 0
|

|
| 11) |
Elute the bisPMP labeled O-glycans with 200 μL of 50% acetonitrile in Milli-Q water and evaporated in a SpeedVac for 1 h at 40°C and resolve in 10 μL of water.
For MALDI-TOF MS analysis, jump to Step 4). |
Comment 0
|
|
|
|
3. |
Sample preparation of formerly glycosylated peptide prior to MALDI-TOF MS analysis |
| 1) |
Add a 4-fold volume of cold ethanol and incubate for 3 h at −30oC. |
Comment 0
|

|
| 2) |
Centrifugate at 20,000 × g for 10 min at 4°C. |
Comment 0
|

|
| 3) |
Wash the pellet with cold ethanol. |
Comment 0
|

|
| 4) |
Digest the pellet with trypsin (or other suitable protease).
For MALDI-TOF MS analysis, jump to Step 4). |
Comment 0
|
|
|
|
4. |
|
| 1) |
Mix each 1 μL of a sample solution and matrix solution and transfer onto a target plate (polished steel plate, 2 mm diameter) and allow to dry. |
Comment 1
|

|
| 2) |
Place the sample plate in the target holder, transfer it into the instrument and wait for the vacuum to form. |
Comment 0
|

|
| 3) |
Select appropriate instrument settings. These settings include the polarity (reflector positive), detected m/z range (500 to 4000), laser power (20–30%), number of summed laser shots (1000). All parameters can be adjusted to maximize the MS data quality. |
Comment 0
|
|
|
| Initial amount | Glycoprotein: ~1 μg
Cells: ~1×106 |
| Discussion | A novel method for the analysis of O-glycosylation of proteins, which occurs either serine or threonine as posttranslational modifications, is described. BEP allows simultaneous release and labeling of O-glycans with pyrazolone analogs without any significant (less than 3%) side reaction (peeling). Under the optimized BEP reaction conditions, the NaOH concentration is as high as ∼180 mM, whereas the presence of the pyrazolone derivative at∼200mM lowers the pH of the reaction solvent to below 9.0, such that the conditions for β-elimination are milder. It should be noted that the BEP procedure is not capable to cleave N-glycans. It is highly advantageous considering that O-glycans and N-glycans may have the same molecular weight and are sometimes difficult to differentiate.
Since pyrazolone is a carbon-carbon bond-forming Michael donor, peptides formerly modified with phosphoric and various O-glycans are concomitantly labeled with the same reagent, thus BEP is unique in that it allows concomitant labeling of both the released O-glycan and the deglycosylated peptide. The ability to label both O-glycan and peptide moieties upon β-elimination is particularly useful in the discrimination between phosphorylation and O-GlcNAcylation. The selectivity of β-elimination toward O-GlcNAcylated and phosphorylated peptides has yet to be addressed properly, such that it is difficult to discriminate between O-GlcNAcylation and phosphorylation by only the β-elimination method. Since BEP allows the detection of both the peptide and the GlcNAc derived from parent glycopeptide/glycoprotein, the reaction can be applied to directly discriminate between GlcNacylated and phosphorylated peptide/protein by detecting the GlcNAc residue.
We examined BEP for phosphorylated and various types of O-glycosylated peptides/proteins and confirmed that BEP is generally applicable to all Ser/Thr modifications so far tested. |
| Figure & Legends |
Figure & Legends

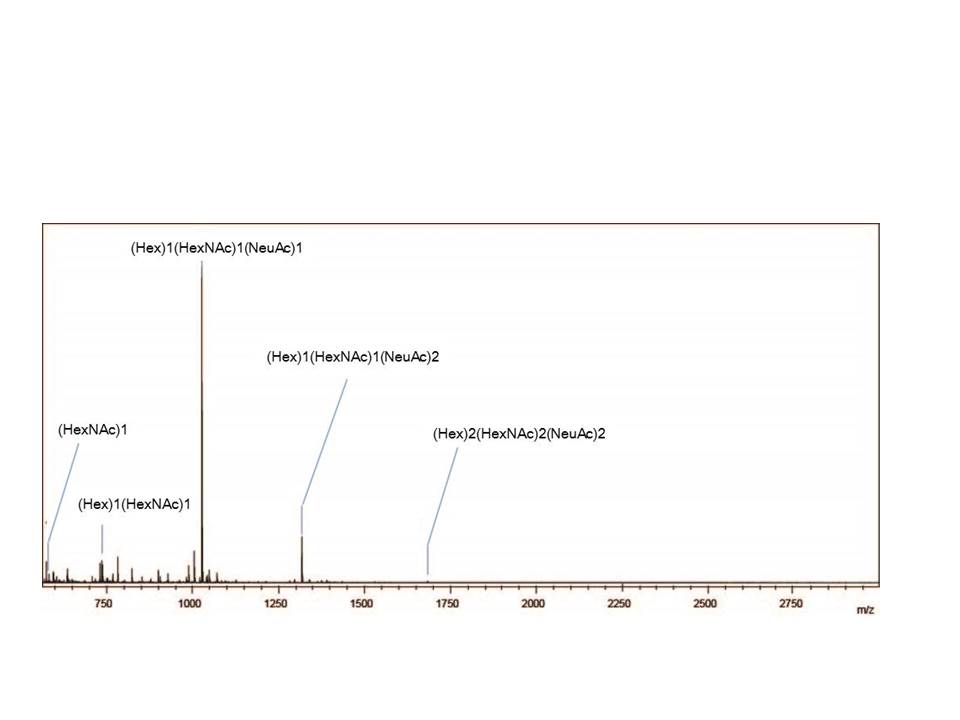
Fig. 1. MALDI-TOF MS spectrum showing the O-glycan profiles of bovine fetuin.

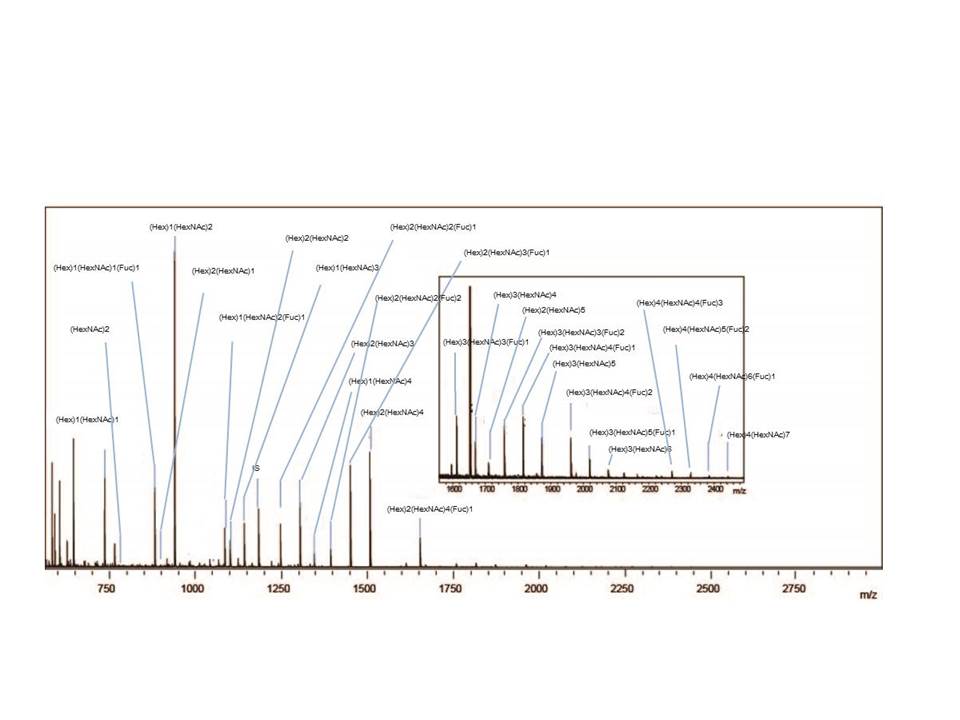
Fig. 2. MALDI-TOF MS spectrum showing the O-glycan profiles of porcine stomach mucin.

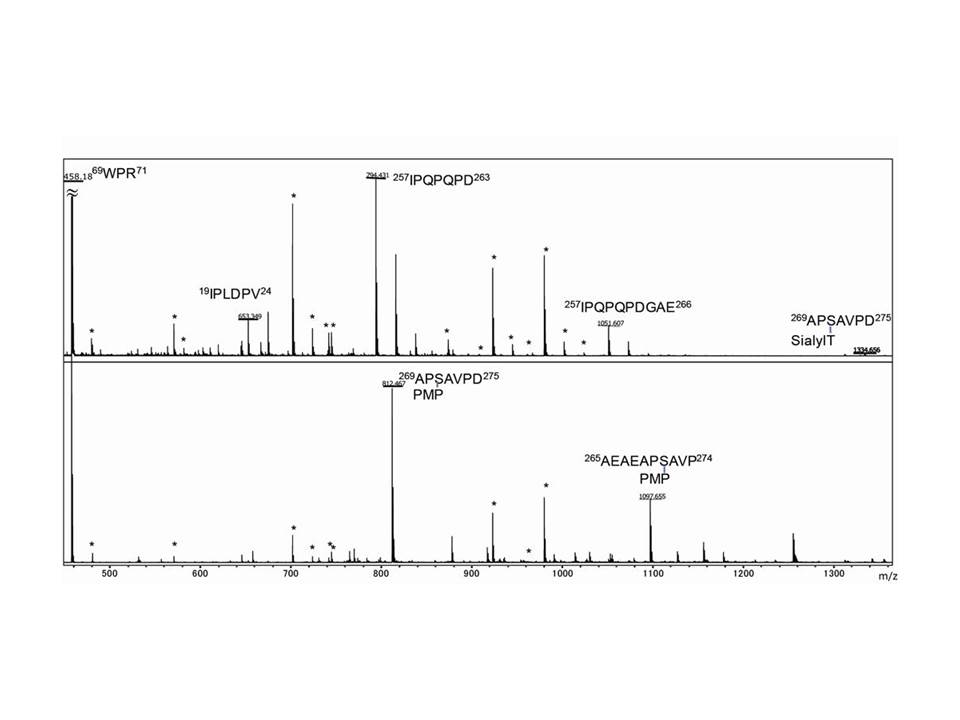
Fig 3. MALDI-TOF MS spectra showing the identification of O-glycosylation sites of bovine fetuin.
Either S-alkylated fetuin (upper panel) or S-alkylated and BEP-treated fetuin (lower panel) was subjected to pronase digestion. Signals attributable to autodigestion of pronase are labeled with asterisk.
These three figure were originally published in "A Versatile Method for Analysis of Serine/Threonine Posttranslational Modifications by β-Elimination in the Presence of Pyrazolone Analogues" Furukawa, J. et al [Anal. Chem. 83 (23), 9060–9067] 2011. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-02-16 15:52:59 |
- Furukawa, J.-i., Fujitani, N., Araki, K., Takegawa, Y., Kodama, K., and Shinohara, Y. (2011) A versatile method for analysis of serine/threonine posttranslational modifications by β-elimination in the presence of pyrazolone analogues. Anal Chem. 83, 9060–9067 [PMID : 21995958]
- Fujitania,N., Furukawa, J.-i., Araki, K., Fujioka, T., Takegawa, Y., Piao, J., Nishioka, T., Tamura, T., Nikaido, T., Ito, M., Nakamura, Y., and Shinohara, Y. (2013) Total cellular glycomics allows characterizing cells and streamlining the discovery process for cellular biomarkers Proc Natl Acad Sci USA. 110, 2105–2110 [PMID : 23345451]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Furukawa, Jun-ichi,
Shinohara, Yasuro,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.18,4,2024 .
How to Cite this Work in Website:
Furukawa, Jun-ichi,
Shinohara, Yasuro,
(2015).
O-glycosylation analysis by β-elimination in the presence of pyrazolone analogue (BEP).
Retrieved 18,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t237.
html source
Furukawa, Jun-ichi,
Shinohara, Yasuro,
(2015).
<b>O-glycosylation analysis by β-elimination in the presence of pyrazolone analogue (BEP)</b>.
Retrieved 4 18,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t237" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t237</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|