Nuclear magnetic resonance (NMR) spectroscopy is a valuable tool to characterize the glycans. Since NMR experiment is non-invasive (non-destructive), it can be considered first among the analytical methods after isolation or preparation of the sample. NMR can offer information on 1) configuration of sugar residues (e.g. Man or Glc), 2) anomeric configuration (α or β), 3) glycosidic linkages, 4) the number of sugar residues, and 5) position of substituents (sulfates, phosphates etc.) 1). The limitation is that the NMR analysis typically requires at least a few nanomoles of the glycan sample. It should be noted that the requisite amount is dependent on the information we need. |
| Category | Isolation & structural analysis of glycans |
| Protocol Name | Determination of glycan structure by NMR |
Authors
 |
Yamaguchi, Yoshiki
Structural Glycobiology Team, RIKEN
|
| KeyWords |
|
| Methods |
|
1. |
Preparation of the glycan sample: |
| 1) |
Isolate the glycan at >90% homogeneity and lyophilize the sample. Since buffers or salts may prevent the NMR analyses, they should be removed in advance as much as possible. |
Comment 0
|

|
| 2) |
Dissolve the sample with 200 or 500 μL of D2O. Transfer the solution into a 5-mm NMR tube (Fig. 1). |
Comment 0
|
|
|
|
2. |
|
| 1) |
Measure the one-dimensional 1H-NMR spectrum (Fig. 2). Identify the “structural reporter group” such as anomeric protons 2). Analyze the chemical shift and vicinal coupling constant (e.g. 3JH1, H2) of each proton signal. |
Comment 0
|

|
| 2) |
If necessary, measure a series of two-dimensional NMR spectra. One-dimensional 13C-NMR typically requires a large amount of sample (10 mg). |
Comment 0
|
|
|
| Notes | *Glycolipid can be dissolved in DMSO-d6 or a mixed solvent (DMSO-d6:D2O = 98:2) 3).
*If a cryogenic probe is used with optimized experimental conditions, picomole amounts of oligosaccharides can be characterized by 1H-NMR spectroscopy 4). |
| Figure & Legends |
Figure & Legends

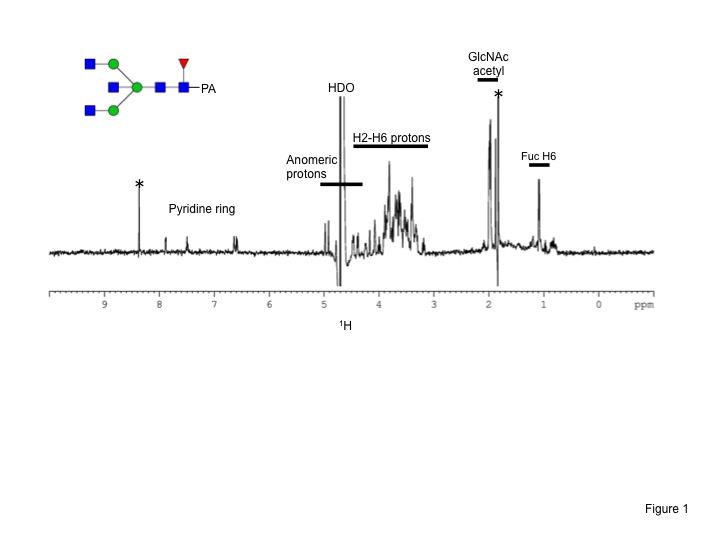
Fig. 1. Standard sample tubes for NMR measurements.
Normal 5-mm NMR tube filled with 500 μL solution (bottom) and Shigemi 5-mm NMR tube (top) with 200 μL solution.

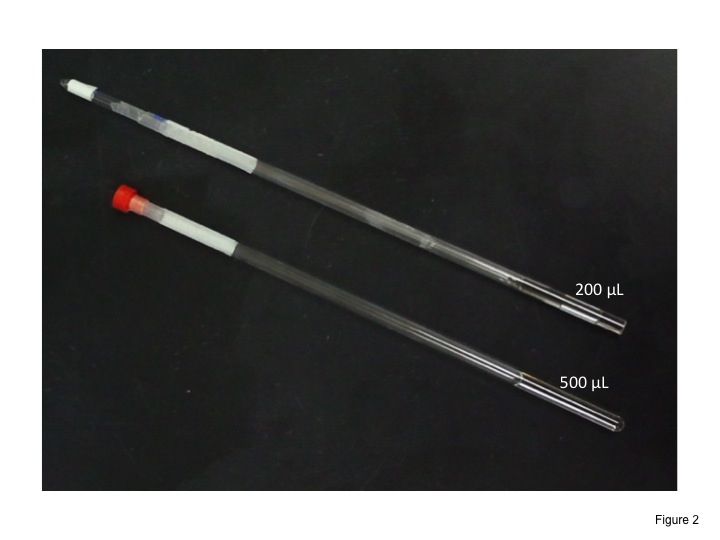
Fig. 2. 500 MHz 1H-NMR spectrum of a pyridylaminated biantennary oligosaccharide dissolved in D2O.
The spectrum was obtained using a DRX-500 spectrometer equipped with aTXI cryogenic probe (BrukerBioSpin). Asterisk indicates the signal from low-molecular-weight impurities. The probe temperature was set at 25°C. 128 scans are acquired and the experimental time is about 5 min. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-02-13 11:33:02 |
- Agrawal, P. K. (1992) NMR spectroscopy in the structural elucidation of oligosaccharides and glycosides. Phytochemistry 31, 3307–3330 [PMID : 1368855]
- Vliegenthart, J. F. G., Dorland, L., and Vanhalbeek, H. (1983) High-resolution, 1H-nuclear magnetic-resonance spectroscopy as a tool in the structural-analysis of carbohydrates related to glycoproteins. Adv. Car. Chem. Biochem. 41, 209–374
- Acquotti, D., and Sonnino, S. (2000) Use of nuclear magnetic resonance spectroscopy in evaluation of ganglioside structure, conformation, and dynamics. Methods Enzymol. 312, 247–272 [PMID : 11070877]
- Fellenberg, M., Çoksezen, A., and Meyer, B. (2010) Characterization of picomole amounts of oligosaccharides from glycoproteins by 1H NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 49, 2630–2633 [PMID : 20198670]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Yamaguchi, Yoshiki,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.19,4,2024 .
How to Cite this Work in Website:
Yamaguchi, Yoshiki,
(2015).
Determination of glycan structure by NMR.
Retrieved 19,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t235.
html source
Yamaguchi, Yoshiki,
(2015).
<b>Determination of glycan structure by NMR</b>.
Retrieved 4 19,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t235" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t235</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|