Methylation is an important tool for the determination of glycan structure. In this method, all the free hydroxyl group of the glycan are methylated and, following hydrolysis of the methylated oligosaccharide, the partially methylated sugars released are reduced by NaBD4 (NaBH4) and acetylated to yield partially methylated alditol acetates (PMAAs). The position of the O-acetyl and O-methyl groups on the PMAA reflects the ring form and linkage positions of the glycosyl residue in the original glycan. PMAAs are usually analyzed by gas chromatography (GC) coupled with electron-impact mass spectrometry (EI-MS). Fragmentation patterns of PMAAs in the mass spectrometer follow well-defined pathways, allowing the mass spectra of these derivatives to be easily recognized (see Fig. 1). This method is widely used for the characterization of a variety of glycoconjugates including polysaccharides and protein-bound glycan (glycoprotein), as well as ceramide-linked glycan (glycosphingolipid, GSL).
Sugar linkage of GSL is characterized by PMAAs derived from permethylated glycosphingolipids. PMAAs obtained after hydrolysis, reduction, and peracetylation are analyzed by the comparison of retention times with authentic standard sugars using GC and confirmed with their mass spectra using gas chromatography / mass spectrometry (GC/MS). |
| Category | Glycolipids and related compounds |
| Protocol Name | Sugar linkage analysis by permethylation (glycosphingolipid) |
Authors
 |
Itonori, Saki
Department of Chemistry, Faculty of Education, Shiga University
|
| KeyWords |
|
Reagents
 |
| ● |
Dimethyl sulfoxide (Nacalai Tesque Inc., Kyoto, Japan) |
| ● |
Iodomethane (Nacalai Tesque Inc.) |
| ● |
Sodium hydroxide (Nacalai Tesque, Kyoto, Japan) |
| ● |
Hydrochloric acid (Nacalai Tesque Inc.) |
| ● |
Glacial acetic acid (Nacalai Tesque Inc.) |
| ● |
Toluene (Nacalai Tesque Inc.) |
| ● |
Sodium borohydride (NaBH4) (Nacalai Tesque Inc.) |
| ● |
Sodium borodeuteride (Sigma-Aldrich, St. Louis, MO) |
| ● |
Pyridine (Nacalai Tesque Inc.) |
| ● |
Acetic anhydride (Nacalai Tesque Inc.) |
| ● |
3M-methanolic HCl (prepare in our laboratory, or Sigma-Aldrich) |
| ● |
n-hexane (Nacalai Tesque Inc.) |
| ● |
Silver carbonate (Nacalai Tesque Inc.) |
| ● |
TMS reagent (prepare in our laboratory; hexamethyldisilazane : trimethylchlorosilane : pyridine = 3:1:9, or Sigma-Aldrich) |
| ● |
Dichloromethane (Nacalai Tesque Inc.) |
| ● |
Boron tribromide (Nacalai Tesque Inc.) |
| ● |
Chloroform (distilled in our laboratoty, Nacalai Tesque Inc.) |
| ● |
Methanol (distilled in our laboratoty, Nacalai Tesque Inc.) |
|
Instruments
 |
| ● |
Round bottom screw glass tube with a PTFE screw cap |
| ● |
|
| ● |
|
| ● |
|
| ● |
N2 evaporator (Do not recommend to use speed vac for PMAA, very volatile) |
| ● |
|
| ● |
Gas chromatography / mass spectrometer (GC/MS) |
|
| Methods |
|
1. |
Permethylation of glycosphingolipid (100 μg – 300 μg) |
| 1) |
Transfer an aliquot of glycolipid solution to a round bottom screw glass tube with a PTFE screw cap and evaporate to dryness. |
Comment 0
|

|
| 2) |
Add 0.2 mL of dimethyl sulfoxide and dissolved into it. |
Comment 0
|

|
| 4) |
Add about 20 mg of powdered sodium hydoride and 0.2 mL of Iodomethane. |
Comment 0
|

|
| 5) |
Immediately capped and vigorously shaken at high speed using a vortex mixer at room temperature for 3 min. |
Comment 0
|

|
| 6) |
Place the tube on the ice and add 1 mL of chloroform and 7 mL of water. |
Comment 0
|

|
| 7) |
Shake with a vortex mixer and then centrifuge at 3,000 rpm. |
Comment 0
|

|
| 8) |
Discard water phase and the add 7 mL of water. |
Comment 0
|

|
| 10) |
Transfer chloroform phase to another tube and dry up. [Permethylated glycolipid] |
Comment 0
|
|
|
|
2. |
Partially methylated alditol acetates from the permethylated glycolipid |
| 1) |
Add 0.3 mL of a mixture of acetic acid, hydrochloric acid, water (8.0:0.5:1.5, v/v). |
Comment 0
|

|
| 3) |
Evaporate under N2 stream with several drops of toluene. |
Comment 0
|

|
| 4) |
Dry up in a desiccator at least for 1h. |
Comment 0
|

|
| 5) |
Add 0.25 mL of 0.01M- sodium hydride and 0.25 mL of 2 % NaBH4 in 0.01M- sodium hydride and incubate at room temperature overnight. |
Comment 0
|

|
| 6) |
Add two or three drops of acetic acid and methanol. |
Comment 0
|

|
| 8) |
Add 0.3mL of 10% AcOH in MeOH and dry under N2 stream (3 times). |
Comment 0
|

|
| 10) |
Add 0.25 mL of pyridine and 0.25 mL of acetic anhydride and incubate in a water bath at 100°C for 10 min. |
Comment 0
|

|
| 11) |
Add 1 mL of chloroform and 4 mL of water. |
Comment 0
|

|
| 12) |
Wash with water 4 times same as above. |
Comment 0
|

|
| 13) |
Transfer chloroform phase to another tube and dry up. [Partially methylated alditol acetates] |
Comment 0
|

|
| 14) |
Analyze the partially methylated alditol acetates by GC and GC/MS. |
Comment 0
|
|
|
|
3. |
Methanolysis of glycosphingolipid (50 μg – 200 μg) |
| 1) |
Transfer an aliquot of glycolipid solution to the one-end rounded glass tube and evaporate to dryness.
(For alternative method, see Comment.) |
Comment 1
|

|
| 2) |
Add 0.2 mL of 1M-methanolic HCl and seal another end of glass tube. [Caution: burst or leak]
(For alternative method, see Comment.) |
Comment 1
|

|
| 3) |
Incubate at 100°C for 3 h in boiling water bath.
(For alternative method, see Comment.) |
Comment 1
|

|
| 4) |
After cooling down, add 0.2 mL of n-hexane and vortex for extraction of fatty acid methyl ester. |
Comment 0
|

|
| 5) |
Flush centrifuge and remove upper hexane phase. |
Comment 0
|

|
| 6) |
Repeat 2 times for extraction of fatty acid methyl ester. |
Comment 0
|

|
| 7) |
Add about 20 mg of silver carbonate to lower phase for neutralization. |
Comment 0
|

|
| 8) |
Remove the reaction salts and excess silver carbonate by small scale filtration. |
Comment 0
|

|
| 9) |
Evaporate to dryness the methanol phase including methyl-glycosides. |
Comment 0
|

|
| 10) |
Add 0.5 mL of methanol, 10 μL of pyridine, and 50 μL of acetic anhydride. |
Comment 0
|

|
| 11) |
Incubate at room temperature for 30 min and then evaporate to dryness. |
Comment 0
|

|
| 12) |
Quickly add about 100 μL of TMS-reagent and incubate at 60°C for 30 min. |
Comment 0
|

|
| 13) |
Add 1 mL of chloroform and 4 mL of water. |
Comment 0
|

|
| 14) |
Wash with water 4 times same as above. |
Comment 0
|

|
| 15) |
Transfer chloroform phase to another tube and dry up. |
Comment 0
|

|
| 16) |
Analyze the trimethylsilyl methyl glycosides by GC and GC/MS. [O-TMS derivatives] |
Comment 0
|
|
|
|
4. |
Boron tribromide demethylation for determination of backbone sugar. |
| 1) |
Prepare methyl glycoside containing O-methylated sugar by methanolysis. |
Comment 0
|

|
| 2) |
Add 0.5 mL of dry dichloromethane and dissolved into it. |
Comment 0
|

|
| 3) |
Place the glass tube at -80°C in a bath of acetone- dry ice. |
Comment 0
|

|
| 4) |
Add 0.2 mL of boron tribromide and maintain at -80°C for 30 min. |
Comment 0
|

|
| 5) |
Allow to regain 20°C, and keep overnight. |
Comment 0
|

|
| 6) |
Remove solvent and excess reagent by evaporation under reduced pressure. |
Comment 0
|

|
| 7) |
Re-methanolysis and prepare O-TMS derivatives for analyze by GC. |
Comment 0
|
|
|
| Notes | 1) Methylation analysis provides only partial information about the linkage position on sugar sequence. Complete structures should be built up with additional analytical results by MS/MS or PSD spectra from MALDI-TOF/MS, LSI/MS, and FAB/MS that support those sugar sequences. Sugar linkage can be analyzed by 2D-NMR with HOHAHA method etc., although it requires large amount of sample (above 1 mg). Therefore sugar linkage analysis by PMAAs is an efficient method for the structural elucidation for small scale biological samples.
2) Some GSLs especially in lower animal are bearing methyl-sugars, e.g. Me-Fuc, Me-Gal, Me-GalNAc, and Me-Xyl. Determination of the position of methyl group on those methylated sugars requires additional analytical approaches as follows: i) TMS derivative for comparison retention time and peak pattern to authentic or chemical synthesized standard sugar by GC, ii) de-methylation by boron tribromide for backbone sugar determination, and iii) alditol acetates derivative for confirmation of mass fragmentation pattern by GC/MS.
3) Mass spectra of partially methylated alditol acetates are available from the superior database in the University of Georgia, Complex Carbohydrate Research Center.
Author thanks Dr. Kazuhiro Aoki of CCRC for his valuable comments. |
| Initial amount | 1. Permethylation of glycosphingolipid (100 μg – 300 μg)
3. Methanolysis of glycosphingolipid (50 μg – 200 μg) |
| Figure & Legends |
Figure & Legends

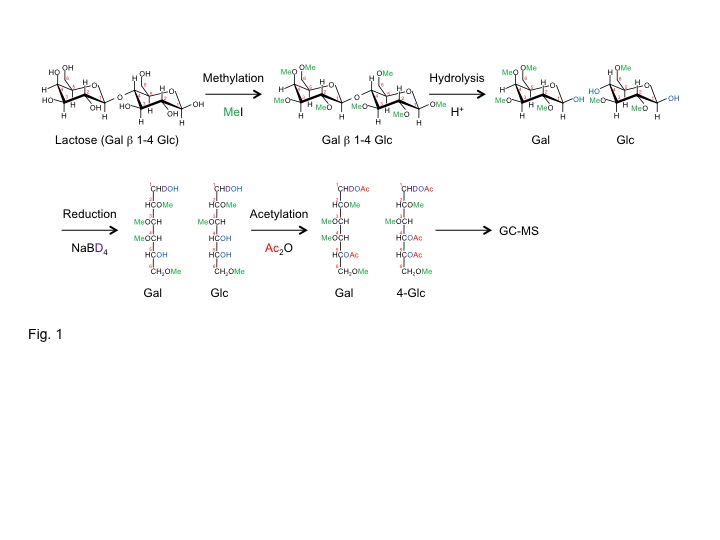
Fig. 1. Sugar linkage analysis by permethylation
Sugar linkage analysis by permethylation
At the first step of permethylation, all free hydroxyl groups in the sugar are protected by persistent blocking with methyl group. Acid hydrolysis makes hydroxyl groups appear at only glycosidic linkages. For analytical purpose, sugar ring structures are opened by reduction with NaBD4 or H4. At the final chemical reaction step, the exposed hydroxyl groups involved in ring and linkage position are acetylated. The resulting volatile sugar derivatives are analyzed by gas chromatography coupled to mass spectrometry. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2014-04-24 16:28:54 |
- Ciucanu, I., and Kerek, F. (1984) A simple and rapid method for the permethylation of carbohydrates. Carbohydr. Res., 131, 209–217.
- Geyer, R., and Geyer, H. (1994) Saccharide linkage analysis using methylation and other techniques. Methods in enzymology 230, 86-108[PMID : 8139517]
- Levery, S., and Hakomori, S. (1987) Microscale methylation analysis of glycolipids using capillary gas chromatography-chemical ionization mass fragmentography with selected ion monitoring. Methods in enzymology 138, 13-25
- Kameyama, A. in this section. “Permethylation for glycan analysis”
- Sweeley, C., Bently, R., Makita, M., and Wells, W. (1963) Gas-liquid chromatography of trimethylsilyl derivatives of sugars and related substances. J. Am. Chem. Soc., 85, 2497-2507.
- Hough, L. and Theobald, R. (1963) Dealkylation. Methods in Carbohydrate Chemistry, II, 203-206.
- Morelle, W. and Michalski, J-C. (2007) Analysis of protein glycosylation by mass spectrometry. Nature protocols, 2, 1585-1602 [PMID : 17585300]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Itonori, Saki,
(2014). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.19,4,2024 .
How to Cite this Work in Website:
Itonori, Saki,
(2014).
Sugar linkage analysis by permethylation (glycosphingolipid).
Retrieved 19,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t225.
html source
Itonori, Saki,
(2014).
<b>Sugar linkage analysis by permethylation (glycosphingolipid)</b>.
Retrieved 4 19,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t225" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t225</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|