Protein glycosylation is one of the major post-translational modifications in eukaryotes, and plays important roles in controlling various cellular processes. We considered that the first step to elucidate the glycan function is to determine actually glycosylated sites on each glycoprotein, and developed a method to determine the glycosylated sites in a large-scale and high-throughput manner. We describe here a protocol for isotope-coded glycosylation site–specific tagging (IGOT), a method for the large-scale identification of N-linked glycoproteins from complex biological sample (Fig.1). The steps of this approach are: (1) affinity capture of glycopeptides with lectin column or hydrophilic interaction chromatography (HILIC) from a protease-digest of protein mixture; (2) purification of the lectin-enriched glycopeptides by HILIC; (3) peptide-N-glycanase-mediated incorporation of a stable isotope tag, 18O, specifically at the N-glycosylation site; and (4) identification of 18O-tagged peptides by liquid chromatography–coupled mass spectrometry (LC/MS)-based proteomics technology. The application of this protocol to the characterization of N-linked glycoproteins from crude extracts of the nematode Caenorhabditis elegans or mouse tissues provides a list of hundreds to a thousand glycoproteins and their sites of glycosylation1)-10). The identified glycoproteins, their glycosylated sites, and the methods for preparation of the glycopeptides (tissues and lectins used) are publically available via our glycoprotein database named GlycoProtDB (http://jcggdb.jp/rcmg/gpdb/index). |
| Category | N-Glycans |
| Protocol Name | Large-scale identification of N-glycosylated peptides using lectin-mediated affinity capture, glycosylation site–specific stable isotope tagging, and LC/MS. |
Authors
 |
Kaji, Hiroyuki
Glycoscience and Glycotechnology Research Group, Biotechnology Research Institute for Drug Discovery, Department of Life Science and Biotechnology, National Institute of Advanced Industrial Science and Technology (AIST)
|
| KeyWords |
|
Reagents
 |
| ● |
Human serum albumin (Sigma-Aldrich, St. Louis, MO) |
| ● |
Guanidine hydrochloride (guanidine-HCl, reagent grade) (see Note 1) |
| ● |
|
| ● |
Ethylenediamine tetraacetic acid·2Na (EDTA) |
| ● |
|
| ● |
2-[4-(2-Hydroxyethyl)-1-piperazinyl]ethanesulfonic acid (HEPES) |
| ● |
Phenylmethylsulphonyl fluoride (PMSF; Sigma-Aldrich) |
| ● |
TPCK-treated trypsin (Thermo Fisher Scientific Inc., Waltham, MA) |
| ● |
Lys-C protease (Achromobacter protease I, Lysyl Endopeptidase; Wako Pure Chemical Industries Ltd., Osaka, Japan) |
| ● |
Methyl α-D-mannopyranoside (for Con A affinity column) |
| ● |
N-Acetylglucosamine (for WGA affinity column) |
| ● |
D-Lactose (for RCA120 and SSA columns) |
| ● |
α-L-Fucose (for AAL column) |
| ● |
Stable isotope, 18O-labeled water (H2 18O, ≥98 atom % 18O; Taiyo Nippon Sanso Corp., Tokyo, Japan) |
| ● |
Peptide-N-glycanase F (PNGase F; Takara Bio Inc., Otsu, Japan) |
| ● |
Acetonitrile (MeCN; chromatography grade, see Note 2) |
| ● |
|
| ● |
Acetic acid (AcOH; see Note 4) |
| ● |
Hydrochloric acid (HCl; see Note 5) |
| ● |
|
| ● |
2-Mercaptoethanol (2-ME; see Note 7) |
| ● |
|
| ● |
REAGENT PREPARATION:
* Protein source materials: Whole organisms such as C. elegans and yeast, tissues, cells, organelles and other biological materials, containing at least 1 mg proteins.
* Extraction buffer: 0.5 M Tris-HCl, pH 8.0, containing 7 M guanidine-HCl and 50 mM EDTA.
* HEPES buffer: 10 mM HEPES-NaOH, pH 7.5.
* Con A binding buffer: 10 mM HEPES-NaOH, pH 7.5, containing 1 mM CaCl2 and 1 mM MnCl2.
* Con A elution buffer: 10 mM HEPES-NaOH, pH 7.5, containing 0.2 M methyl α-D-mannopyranoside.
* Hydrophilic interaction chromatography (HILIC) solvent A: 50 % acetonitrile (MeCN, v/v) containing 0.1 % TFA (v/v).
* Hydrophilic interaction chromatography (HILIC) solvent B: 75 % MeCN containing 0.1 % TFA.
* PNGase buffer for stable isotope-tagging: 0.1 M Tris-acetic acid (AcOH), pH 8.6 (Dissolve Tris base in H218O and adjust at pH 8.6 with 1 M AcOH in H218O).
* Reverse-phase column solvent A: 0.1% (v/v) formic acid in water.
* Reverse-phase column solvent B: 0.1% (v/v) formic acid in acetonitrile. |
|
Instruments
 |
| ● |
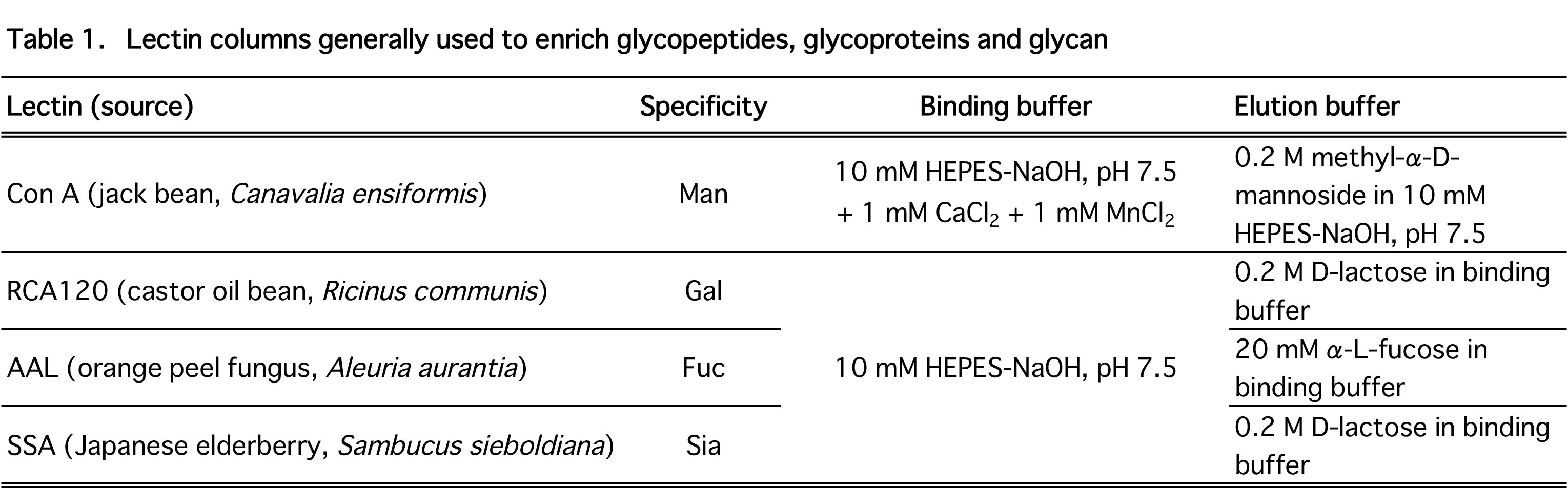
Lectin-affinity column (e.g., Con A column; Table 1), 4.6 mm × 150 mm for high performance liquid chromatography (HPLC) (J-OIL MILLS, Inc., Tokyo, Japan) |
| ● |
Amide-80 column, 2 mm × 50 mm (Tosoh Corp., Tokyo, Japan) |
| ● |
Reverse-phase column, 0.15 mm × 50 mm (packed in-house with Mightysil RP 18 GP, 3 μM particles; Cica) or 0.075 mm × 100 mm (Nikkyo Technos Co., Ltd., Tokyo, Japan) |
| ● |
Nanoflow HPLC system (LC-Assist) |
| ● |
Centrifugal vacuum concentrator |
| ● |
Electrospray-ionization high resolution tandem mass spectrometer (e.g., LTQ-Orbitrap MS, Thermo Fisher Scientific Inc., Waltham, MA) |
| ● |
Mascot (Matrix Science Inc., Boston, MA) |
| ● |
INSTRUMENT SETUP:
* Reverse-phase liquid chromatography Setup a gradient LC with reverse-phase column solvents A and B and set the flow rate to 100 nL min-1. |
|
| Methods |
|
1. |
|
| 1) |
Dissolve the protein source materials (whole organisms, tissues, cells, organelles or other biological materials) in extraction buffer (we usually use 5–10 mL extraction buffer for 1 g material) using the appropriate equipment (e.g., sonicator, Teflon homogenizer, Waring blender, French press, Polytron). Remove insoluble materials, if any, by centrifugation at 10,000g for 30 min at room temperature (20°C). |
Comment 0
|

|
| 2) |
Quantify protein concentration by Bradford assay using human serum albumin as a standard. |
Comment 0
|

|
| 3) |
Add an aliquot of 100 mg mL-1 DTT to the sample solution at DTT:protein = 1:1 (w/w), and incubate with gentle shaking for 2 h at room temperature to reduce disulfide bonds under nitrogen gas atmosphere. For a small-volume sample, perform the reduction in a sealed tube under N2 gas. |
Comment 0
|

|
| 4) |
For S-carbamoylmethylation of cysteine residues, add iodoacetamide to the protein solution (2.5:1 w/w protein), mix and leave the mixture in the dark for 2 h at room temperature. |
Comment 0
|

|
| 5) |
Dialyze the reaction mixture at 4°C against 100 volumes of 10 mM HEPES-NaOH buffer, pH 7.5, to remove salts and excess reagents, changing the buffer three times for every 2 h and then leave overnight. For a small-volume sample, precipitate the protein by acetone, trichloroacetic acid, or methanol/chloroform precipitation method. |
Comment 0
|

|
| 6) |
Transfer the dialyzed mixture to a plastic conical tube. Add trypsin (1:50 w/w protein) or Lys-C protease (1:100 w/w protein) to the solution and digest the proteins overnight at 37°C (see Comment). |
Comment 1
|

|
| 7) |
Add an aliquot of 0.5 M PMSF in EtOH into the mixture at a final concentration of 1 mM to stop digestion and to protect the lectin column. Remove precipitates, if any, by centrifugation. |
Comment 0
|
|
|
|
2. |
Capturing of glycopeptides by lectin-affinity chromatography
To collect glycopeptides comprehensively (regardless of the glycan structure), skip this step to Title 3. |
| 1) |
Load the peptide mixture onto the lectin column equilibrated with a buffer appropriate for each lectin column (listed in Table 1). Take care not to overload the column — a typical lectin column with 4.6 mM ID × 150 mM can separate - 400 μg glycopeptides. Recover the flow-through fraction. |
Comment 0
|

|
| 2) |
Wash the column with the binding buffer until the absorption of the effluent at 280 nm is <0.1 (about 10- column -volumes of buffer is necessary). |
Comment 0
|

|
| 3) |
Elute glycopeptides with an elution buffer containing a mono/oligosaccharide appropriate for the lectin column (e.g., 0.2 M methyl α-D-mannopyranoside for a Con A column, see Table 1). |
Comment 0
|

|
| 4) |
Re-equilibrate the lectin column with five -column- volumes of binding buffer. |
Comment 0
|

|
| 5) |
Reload the flow-through fraction onto the lectin column and recollect the glycopeptide fraction under the same conditions as above. Repeat the chromatography three to five times until the absorption of the glycopeptide peak at 280 nm is <0.1. Combine the glycopeptide fractions. |
Comment 0
|
|
|
|
3. |
Purification of glycopeptides by HILIC |
| 1) |
Add 3 volumes of MeCN containing 0.1% TFA into the glycopeptide solution eluted from the lectin column (or protease digest). Remove precipitates, if any, by centrifugation (see Comment). |
Comment 1
|

|
| 2) |
Immediately load the sample solution onto an Amide-80 column, which has been equilibrated with HILIC solvent B, and wash the column with the HILIC solvent B until the absorption of the effluent at 220 nm is <0.2. |
Comment 0
|

|
| 3) |
Elute glycopeptides with HILIC solvent A, monitoring the eluate at 220 nm. Samples can be stored at -80°C for several days. |
Comment 0
|
|
|
|
4. |
Glycosylation site–specific stable isotope tagging by PNGase treatment |
| 1) |
Transfer -200 μL of the HILIC eluate into a 500-μL microtube and evaporate to dryness using a centrifugal vacuum concentrator. Add another 200-μL aliquot of the HILIC eluate into the same microtube and evaporate. Repeat the process to dry up the total eluate (usually 1–2 mL) (see Comment). |
Comment 1
|

|
| 2) |
Dissolve glycopeptides in PNGase buffer (10–20 μg/50–100 μL) prepared with H218O. |
Comment 0
|

|
| 3) |
Add PNGase, dissolved in H218O at 1 mU uL–1, at a final concentration of 1 mU 10 μg–1 glycopeptides and incubate the mixture at 37°C overnight in a sealed microtube. |
Comment 0
|

|
| 4) |
Stop the reaction by acidifying the solution to -pH 2 by the addition of 1% formic acid (-5 μL per 100 μL reaction volume). |
Comment 0
|
|
|
|
5. |
LC-MS/MS analysis: Protein identification and determination of the glycosylated site |
| 1) |
Analyze the 18O-labeled peptide mixture using an LC system connected to an electrospray-ionization tandem mass spectrometer. See Comment for details. |
Comment 1
|

|
| 2) |
Search the MS/MS spectra using a nonredundant protein database, such as Refseq and SwissProtKB, using Mascot algorithms for peptide sequence identification with the following parameters: fixed modification of carbamoylmethylation on Cys, variable modifications of deamidation on Asn (plus an incorporation of 18O = +3 Da) and pyroglutamination on N-terminal Gln. |
Comment 0
|

|
| 3) |
Identify the formerly N-glycosylated peptides. Export the result to csv file and process the data with Microsoft Excel. We first selected the peptides with rank 1 and an expectation value <0.05 as “identified peptides”. |
Comment 0
|

|
| 4) |
Inspect the data set to determine whether the 18O-labeled peptide has one or more consensus sequences for N-linked glycosylation (i.e., Asn-Xaa-Ser/Thr, where Xaa is any amino acid except Pro). If the candidate peptide has an Asn-Lys or Asn-Arg sequence at the C terminus, examine if the residue following the Lys/Arg in the protein sequence might be Ser or Thr, which meets the consensus tripeptide sequence. |
Comment 0
|
|
|
| Notes | 1. Harmful if swallowed. Irritating to eyes and skin. Avoid contact with eyes, skin and clothing.
2. Toxic. Wear gloves and use a pipetting aid.
3. Causes severe burns. Wear gloves and use a pipetting aid.
4. Flammable. Causes severe burns. Wear gloves and use a pipetting aid.
5. Toxic if inhaled. Causes severe burns. Wear suitable protective clothing, gloves and eye/face protection.
6. Highly flammable liquid and vapor. Causes eye irritation. May damage fertility or an unborn child. Use only in a well-ventilated area.
7. Harmful if swallowed. Toxic in contact with skin. Causes burns. |
| Initial amount | Preferably >1 mg glycoprotein in a source extract. |
| Produced amount | See anticipated results in Discussion. |
| Discussion | ANTICIPATED RESULTS:
One hundred mg of mouse tissue, e.g., liver or brain (about 10 mg protein or 1 mg of glycoprotein) yields -10–20 μg glycopeptides captured on a Con A column. Processing of the glycopeptides using the protocol described here, followed by identification of 18O-labeled peptides by a single LC-MS/MS analysis, will identify -1,000 N-glycosylated sites on -600 proteins. This largely depends on performance of the LC and MS system. A typical mass spectrum of an 18O-labeled peptide is shown in Fig. 2. |
| Figure & Legends |
Figure & Legends

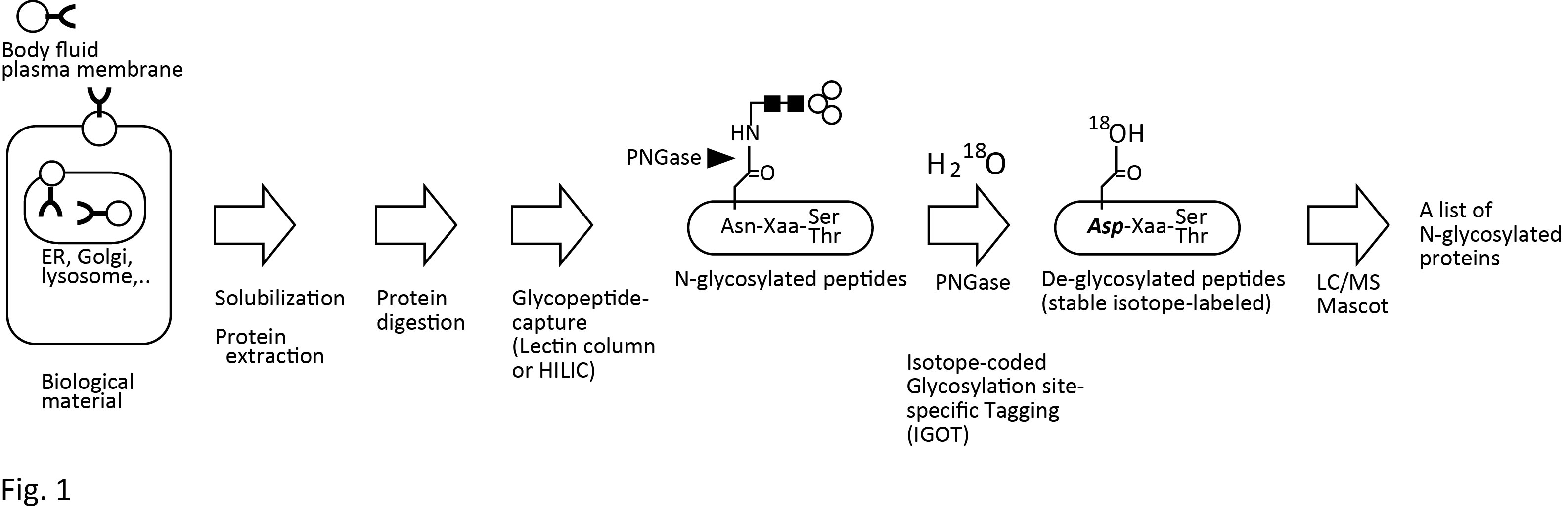
Fig. 1. Outline of IGOT-LC/MS glycoprotein identification method.
See references for detail.

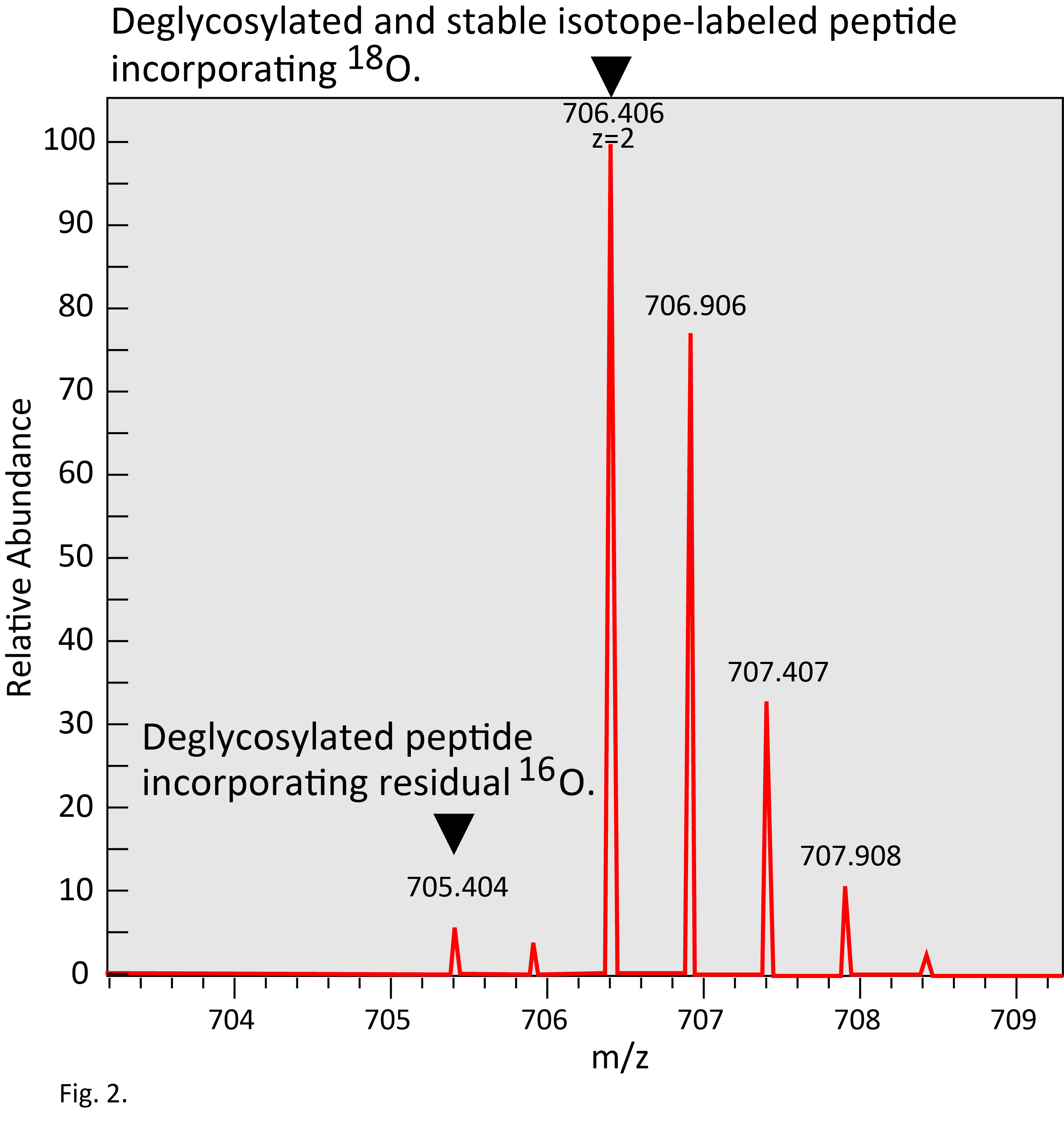
Fig. 2. Typical mass spectrum of IGOT-peptide.
IGOT-peptide shows mass shift of +3 Da from unmodified peptide. Simultaneously, a signal of deglycosylated peptide incorporating ordinary oxygen, 16O, remaining in a solvent is observed.

|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2014-12-16 14:40:08 |
- Kaji, H., Yamauchi Y., Takahashi N., and Isobe, T. (2006) Mass spectrometric identification of N-linked glycopeptides using lectin-mediated affinity capture and glycosylation site–specific stable isotope tagging. Nat Protoc. 1, 3019–27. [PMID : 17406563]
- Kaji H., Saito H., Yamauchi Y., Shinkawa T., Taoka M., Hirabayashi J., Kasai K., Takahashi N., Isobe T. (2003) Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat Biotechnol. 21, 667–72. [PMID : 12754521]
- Natsume, T., Yamauchi, Y., Nakayama, H., Shinkawa, T., Yanagida, M., Takahashi, N., and Isobe, T. (2002) A direct nanoflow liquid chromatography-tandem mass spectrometry system for interaction proteomics. Anal Chem. 74, 4725–33. [PMID : 12349976]
- Isobe, T. et al. (2003) Automated two-dimensional liquid chromatography/tandem mass spectrometry for large-scale protein analysis. In: Proteins and Proteomics, 869–876, Cold Spring Harbor Press, Cold Spring Harbor, NY.
- Mawuenyega, K.G., Kaji, H., Yamauchi, Y., Shinkawa, T., Saito, H., Taoka, M., Takahashi, N., and Isobe, T. (2003) Large-scale identification of Caenorhabditis elegans proteins by multi-dimensional liquid chromatography-tandem mass spectrometry. J Proteome Res. 2, 23–35. [PMID : 12643540]
- Kaji H., Kamiie J., Kawakami H., Kido K., Yamauchi Y., Shinkawa T., Taoka M., Takahashi N., Isobe T. (2007) Proteomics reveals N-linked glycoprotein diversity in Caenorhabditis elegans and suggests an atypical translocation mechanism for integral membrane proteins. Mol Cell Proteomics. 6, 2100–9. [PMID : 17761667]
- Kaji H., Shikanai T., Sasaki-Sawa A, Wen H, Fujita M, Suzuki Y, Sugahara D, Sawaki H, Yamauchi Y, Shinkawa T, Taoka M, Takahashi N, Isobe T, Narimatsu H. (2012) Large-scale identification of N-glycosylated proteins of mouse tissues and construction of a glycoprotein database, GlycoProtDB. J Proteome Res. 11, 4553–66. [PMID : 22823882]
- Kaji H and Isobe T. (2008) Liquid Chromatography/Mass Spectrometry (LC/MS)-Based Glycoproteomics Technologies for Cancer Biomarker Discovery. Clinical Proteomics 2, 14–24
- Sugahara D., Kaji H., Sugihara K., Asano M., Narimatsu H. (2012) Large-scale identification of target proteins of a glycosyltransferase isozyme by Lectin-IGOT-LC/MS, an LC/MS-based glycoproteomic approach. Sci Rep. 2, 680. [PMID : 23002422]
- Hiroyuki Kaji, Makoto Ocho, Akira Togayachi, Atsushi Kuno, Maki Sogabe, Takashi Ohkura, Hirofumi Nozaki, Takashi Angata, Yasunori Chiba, Hidenori Ozaki, Jun Hirabayashi, Yasuhito Tanaka, Masashi Mizokami, Yuzuru Ikehara, and Hisashi Narimatsu. (2013) Glycoproteomic Discovery of Serological Biomarker Candidates for HCV/HBV Infection-associated Liver Fibrosis and Hepatocellular Carcinoma. J. Proteome Res. 12, 2630–40. [PMID : 23586699]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Kaji, Hiroyuki,
(2014). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.26,4,2024 .
How to Cite this Work in Website:
Kaji, Hiroyuki,
(2014).
Large-scale identification of N-glycosylated peptides using lectin-mediated affinity capture, glycosylation site–specific stable isotope tagging, and LC/MS..
Retrieved 26,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t222.
html source
Kaji, Hiroyuki,
(2014).
<b>Large-scale identification of <em>N</em>-glycosylated peptides using lectin-mediated affinity capture, glycosylation site–specific stable isotope tagging, and LC/MS.</b>.
Retrieved 4 26,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t222" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t222</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|