The endoplasmic reticulum (ER) is the organelle in which secretory and membrane proteins are synthesized. Most of the polypeptides synthesized in the ER are covalently modified with N-linked oligosaccharides. The folding of these glycoproteins is monitored and assisted by chaperone proteins and lectins in the ER. Correctly folded proteins are sorted out of the ER into secretory compartments, whereas misfolded polypeptides or unassembled subunits are retained in the ER. After several attempts to correctly fold these polypeptides, terminally misfolded proteins are retro-translocated out of the ER and degraded by the cytoplasmic proteasome. This mechanism is termed ER-associated degradation (ERAD) 1)-3).
Many steps are required for ERAD including recognition of misfolded ERAD substrates, extraction of these proteins from the ER to the cytosol, polyubiquitination, deglycosylation and finally protein degradation by the proteasome. To analyze the function of molecules involved in these processes, the degradation of ERAD substrates transfected into mammalian cells is monitored. |
| Category | Biosynthesis & Metabolism |
| Protocol Name | Assay for endoplasmic reticulum-associated degradation (ERAD) in mammalian cells |
Authors
 |
Hosokawa, Nobuko
Department of Molecular and Cellular Biology, Institute for Frontier Medical Sciences, Kyoto University
|
| KeyWords |
|
Reagents
 |
| ● |
Transfection reagents: Lipofectamine 2000 (Invitrogen/Life Technologies, Carlsbad, CA), Effecten (Qiagen, Venlo, Netherlands), and others |
| ● |
[35S]-methionine/cysteine (or [35S]-protein labeling mixture) |
| ● |
DMEM lacking methionine/cysteine |
| ● |
Protein A- or Protein G-Sepharose beads (GE Healthcare, Little Chalfont, UK) |
| ● |
|
| ● |
Proteasome inhibitors: lactacystin, MG-132, epoxomicin and others |
| ● |
Imaging plate : Fujifilm BAS-TR Imaging Plates (GE Healthcare) and others |
| ● |
LuminoImage analyzer: LAS 4000 (GE Healthcare) and others |
|
Instruments
 |
| ● |
CO2 incubator (Panasonic, Osaka, Japan, Napco/Thermo Fisher Scientific Inc., Waltham, MA, TAITEC Co., Ltd., Koshigaya, Japan and others) |
| ● |
Radioisotope experiment facility |
| ● |
Rotator (As One Corporation, Osaka, Japan, TAITEC Co., Ltd. and others) |
| ● |
PhosphorImager : Typhooon (GE Healthcare), BAS (GE Healthcare) |
|
| Methods |
|
1. |
|
| 1) |
Plate cells on a 3.5 cm tissue culture dish. |
Comment 1
|

|
| 2) |
Transfect plasmids encoding an ERAD substrate and proteins of interest. |
Comment 1
|

|
| 3) |
Incubate cells in DMEM lacking methionine/cysteine for 15–30 min. |
Comment 0
|

|
| 4) |
Pulse-label cells with [35S]-methionine/cysteine for 15 min in DMEM lacking methionine/cysteine. |
Comment 1
|

|
| 5) |
Remove medium and incubate cells in normal growth medium for the time required. |
Comment 0
|

|
| 6) |
Harvest and extract cells in a buffer containing an appropriate detergent. |
Comment 1
|

|
| 7) |
Add antibodies to cell lysates and incubate at 4°C for the time required. |
Comment 1
|

|
| 8) |
Add Protein A- or Protein G-Sepharose beads. |
Comment 1
|

|
| 10) |
Collect beads by centrifugation at 1,500 × g for 3 min at 4°C. |
Comment 0
|

|
| 11) |
Wash beads twice with high ionic buffer and collect beads as in step 10. |
Comment 1
|

|
| 12) |
Wash beads once with low ionic buffer or phosphate buffered saline and collect beads as in step 10. |
Comment 0
|

|
| 13) |
Elute immunoprecipitates with Laemmli sample buffer. |
Comment 0
|

|
| 15) |
Dry the gel and expose to an imaging plate. |
Comment 0
|

|
| 16) |
Analyze using an phosphorImager. |
Comment 0
|
|
|
|
2. |
Cycloheximide-chase experiment |
| 1) |
Plate cells on a 3.5 cm tissue culture dish. |
Comment 0
|

|
| 2) |
Transfect plasmids encoding an ERAD substrate and proteins of interest. |
Comment 0
|

|
| 3) |
Add 100 μg/mL of cycloheximide to the medium. |
Comment 1
|

|
| 4) |
Harvest and extract cells in a buffer containing an appropriate detergent. |
Comment 0
|

|
| 5) |
Add Laemmli sample buffer to cell extracts and separate proteins by SDS-PAGE. |
Comment 0
|

|
| 6) |
Transfer the gel to nitrocellulose or PVDF membrane. |
Comment 0
|

|
| 7) |
Detect specific proteins using appropriate antibodies (Western blotting). |
Comment 0
|
|
|
| Initial amount | Cultured cells: approximately 1 × 106 cells |
| Discussion | A truncation mutant of α1-antytrypsin null Hong Kong (NHK) is a glycoprotein that is degraded by ERAD 4). NHK degradation was enhanced by co-transfection with ER degradation enhancing α-mannosidase-like protein 3 (EDEM3) 5), as observed in a pulse-chase experiment (Fig. 1). NHK degradation was inhibited following the addition of the proteasome inhibitor Lactacystin (Fig. 2), indicating that the proteasome was responsible for this degradation.
SEL1L, the human homolog of C. elegans sel-1, is an ER membrane protein that forms a stoichiometric complex with the ubiquitin ligase, HRD1. Knockdown of endogenous SEL1L by siRNA treatment inhibited NHK degradation (Fig. 3) 6) 7), suggesting that SEL1L is required for the degradation of NHK.
The degradation of ERAD substrates can be quantitatively assessed using these pulse-chase experiments. This method may be useful to identify components involved in ERAD and to assess their roles within the pathway. Although cycloheximide chase experiments are more convenient than pulse-chase experiments, it should be noted that the total amount of protein accumulated in cells is detected by this method, and that not just ERAD substrates but also the synthesis of most cellular proteins is inhibited. Thus, the addition of cycloheximide inhibits the degradation of some ERAD substrates, including NHK 8). |
| Figure & Legends |
Figure & Legends

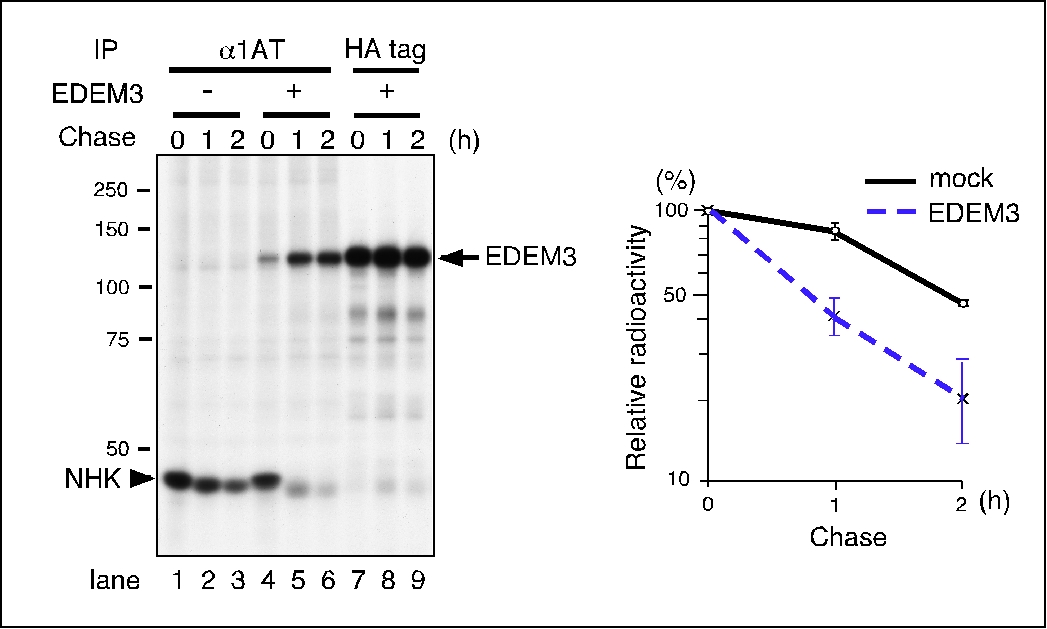
Fig. 1. EDEM3 enhances the degradation of NHK.
HEK (human embryonic kidney) 293 cells were transfected with plasmids encoding NHK, together with an empty vector or EDEM3-HA using FuGENE 6 transfection reagent (Roche), and labeled with [35S]-protein labeling mixture for 15 min. NHK and EDEM3 were immunoprecipitated with an anti- α1-antytrypsin antibody (lanes 1–6) or with an anti-HA-tag antibody (lanes 7–9). Immunoprecipitates were separated on a 10 % SDS-PAGE gel, which was then dried and exposed to an imaging plate. The radioactive signal was quantified using a Typhoon PhosphorImager (GE Healthcare Life Sciences) (right panel).
This figure was originally published in J. Biol. Chem. Hirao K. et al. “EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming” 2006, 281: 9650–9658. © the American Society for Biochemistry and Molecular Biology.

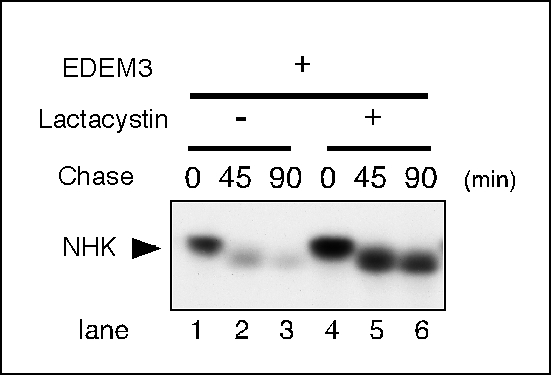
Fig. 2. Lactacystin inhibits the degradation of NHK.
Cells were plated and transfected as described in the legend of Figure 1. Cells were treated with 20 μM lactacystin for 3 h prior to pulse-labeling (lanes 4–6, indicated as +) or were untreated (lanes 1–3, indicated as -).
This figure was originally published in the J. Biol. Chem. Hirao K. et al. “EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming” 2006, 281: 9650–9658/ © the American Society for Biochemistry and Molecular Biology.

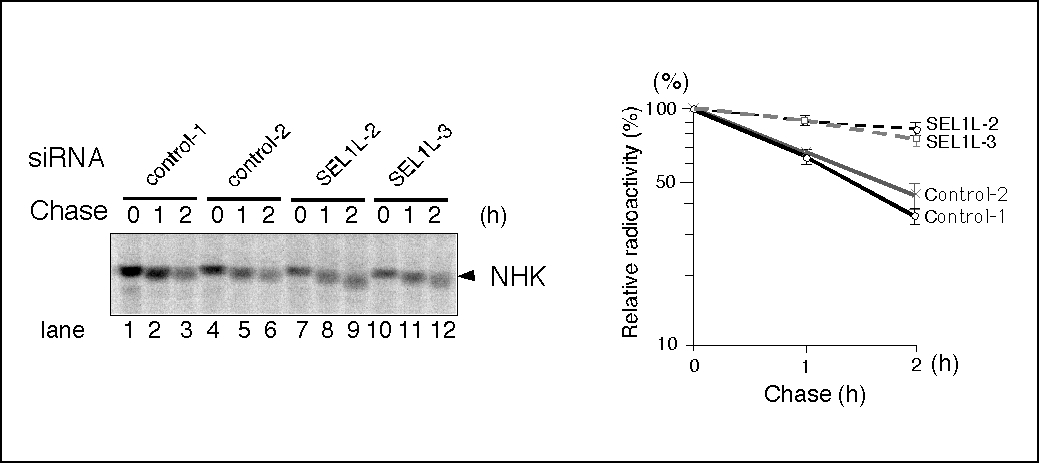
Fig. 3. SEL1L is required for the degradation of NHK.
HEK293 cells were treated with 30 nM of negative control Stealth™ siRNAs (Invitrogen/Life Technologies) (control-1 and -2, lanes 1–6) or specific siRNAs targeting SEL1L (SEL1L-2 and -3, lanes 7-12), and NHK degradation was analyzed as described in the legend of Fig. 1.
This figure was originally published in the J. Biol. Chem. Hosokawa N. et al. “Human XTP3-B Forms an Endoplasmic Reticulum Quality Control Scaffold with the HRD1-SEL1L Ubiquitin Ligase Complex and BiP” 2008, 283: 20914–20924. © the American Society for Biochemistry and Molecular Biology. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2014-09-18 11:37:04 |
- Helenius, A., and Aebi, M. (2001) Intracellular functions of N-linked glycans. Science 291, 2364–2369 [PMID : 11269317]
- Trombetta, E. S., and Parodi, A. J. (2003) Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 19, 649–676 [PMID : 14570585]
- Nakatsukasa, K., and Brodsky, J. L. (2008) The recognition and retrotranslocation of misfolded proteins from the endoplasmic reticulum. Traffic 9, 861–870 [PMID : 18315532]
- Liu, Y., Choudhury, P., Cabral, C. M., and Sifers, R. N. (1999) Oligosaccharide modification in the early secretory pathway directs the selection of a misfolded glycoprotein for degradation by the proteasome. J Biol Chem. 274, 5861–5867 [PMID : 10026209]
- Hirao, K., Natsuka, Y., Tamura, T., Wada, I., Morito, D., Natsuka, S., Romero, P., Sleno, B., Tremblay, L. O., Herscovics, A., Nagata, K., and Hosokawa, N. (2006) EDEM3, a soluble EDEM homolog, enhances glycoprotein endoplasmic reticulum-associated degradation and mannose trimming. J Biol Chem. 281, 9650–9658 [PMID : 16431915]
- Hosokawa, N., Wada, I., Nagasawa, K., Moriyama, T., Okawa, K., and Nagata, K. (2008) Human XTP3-B Forms an Endoplasmic Reticulum Quality Control Scaffold with the HRD1-SEL1L Ubiquitin Ligase Complex and BiP. J Biol Chem. 283, 20914–20924 [PMID : 18502753]
- Christianson, J. C., Shaler, T. A., Tyler, R. E., and Kopito, R. R. (2008) OS-9 and GRP94 deliver mutant alpha1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 10, 272–282 [PMID : 18264092]
- Wu, Y., Termine, D. J., Swulius, M. T., Moremen, K. W., and Sifers, R. N. (2007) Human endoplasmic reticulum mannosidase I is subject to regulated proteolysis. J Biol Chem. 282, 4841–4849 [PMID : 17166854]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Hosokawa, Nobuko,
(2014). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.20,4,2024 .
How to Cite this Work in Website:
Hosokawa, Nobuko,
(2014).
Assay for endoplasmic reticulum-associated degradation (ERAD) in mammalian cells.
Retrieved 20,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t197.
html source
Hosokawa, Nobuko,
(2014).
<b>Assay for endoplasmic reticulum-associated degradation (ERAD) in mammalian cells</b>.
Retrieved 4 20,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t197" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t197</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|