Bacterial lyases are useful for depolymerizing hyaluronan (HA) because of their strong enzymatic activity. Five bacterial glycosaminoglycan lyases able to degrade HA are available commercially (Table 1). Although all of them can cleave HA chains, they each differ in activity and specificity. |
| Category | Glycosaminoglycans |
| Protocol Name | Bacterial glycosaminoglycan lyases to depolymerize hyaluronan |
Authors
 |
Yamada, Shuhei
Department of Pathobiochemistry, Faculty of Pharmacy, Meijo University
|
| KeyWords |
|
Reagents
 |
| ● |
HA preparation having an average molecular mass of 35 kDa produced by microbial fermentation of Streptococcus pyogenes (R & D Systems, Minneapolis, MN) |
| ● |
Hyaluronidase from Streptomyces hyalurolyticus (Seikagaku Biobusiness Corp., Tokyo, Japan) |
| ● |
Hyaluronidase SD from Streptococcus dysgalactiae (Seikagaku Biobusiness Corp., Tokyo, Japan) |
| ● |
Chondroitinase ABC (a conventional preparation) from Proteus vulgaris (Seikagaku Biobusiness Corp., Tokyo, Japan) |
| ● |
Chondroitinase AC-I from Flavobacterim heparinum (Seikagaku Biobusiness Corp., Tokyo, Japan) |
| ● |
Chondroitinase AC-II from Arthrobacter aurescens (Seikagaku Biobusiness Corp., Tokyo, Japan) |
|
Instruments
 |
| ● |
YMC-Pack PA-03 (4.6 mm i.d. x 250mm) (YMC Co., Ltd., Kyoto, Japan) |
| ● |
HPLC system (LC-20 Series; Shimadzu Corp. Kyoto, Japan) |
| ● |
Ultrafree-MC (Merck Millipore, Billerica, MA) |
|
| Methods |
|
1. |
Bacterial glycosaminoglycan lyases to depolymerize hyaluronan |
| 1) |
Enzymatic digestion of HA
a) Incubate HA (2 μg) with hyaluronidase from Streptomyces hyalurolyticus (0.5 TRU) in a total volume of 20 μL of 20 mM acetate-Na buffer, pH 6.0, containing 150 mM sodium chloride at 60˚C for 75 min or hyaluronidase SD from Streptococcus dysgalactiae (5 mIU) in a total volume of 20 μL of 40 mM phosphate-Na buffer, pH 6.2, at 37˚C for 30 min.
b) Incubate HA (2 μg) with chondroitinase (5 mIU each) in a total volume of 20 μL of 50 mM Tris-HCl buffer, pH 8.0, containing 60 mM sodium acetate (chondroitinase ABC), 50 mM Tris-HCl buffer, pH 7.3 (chondroitinase AC-I), or 50 mM acetate-Na buffer, pH 6.0 (chondroitinase AC-II), at 37˚C for 30 min. |
Comment 1
|

|
| 2) |
Terminate the reactions by boiling at 100˚C for 1 min. |
Comment 0
|

|
| 3) |
Dilute the reaction mixture with distilled water to 200 μL. |
Comment 0
|

|
| 4) |
Analyze an aliquot (100 μL) by HPLC on an amine-bound silica PA-03 column using a linear gradient of NaH2PO4 from 16 to 530 mM at a flow rate of 1 mL/min, monitored by measuring UV with a wavelength of 232 nm. |
Comment 0
|

|
| 5) |
Identify and quantify the eluted peaks by comparing with the positions of authentic standards (Fig. 1 and 2) |
Comment 1
|
|
|
| Figure & Legends |
Figure & Legends
| Enzyme |
Substrate Specificity |
Predominant End Products |
|
Hyaluronidase from
Streptomyces hyalurolyticus
|
This enzyme is quite specific for HA, and acts on neither chondroitin sulfate (CS) nor chondroitin.
|
Unsaturated tetrasaccharide and hexasaccharide |
| Hyaluronidase SD from Streptococcus dysgalactiae |
This enzyme acts on not only HA but also chondroitin. However, it does not depolymerize CS. The rate of degradation of chondroitin is much slower than that of HA.
|
Unsaturated disaccharide |
| Chondroitinase ABC from Proteus vulgaris |
HA is hardly degraded by this enzyme. It acts on both CS and chondroitin, but the rate of depolymerization of chondroitin is slower than that of CS. |
Unsaturated disaccharide |
| Chondroitinase AC-I from Flavobacterim heparinum |
This enzyme depolymerizes CS and chondroitin at a similar rate. It also degrades HA, though slightly slower than CS/chondroitin.
|
Unsaturated disaccharide |
| Chondroitinase AC-II from Arthrobacter aurescens |
This enzyme acts on not only CS/chondroitin but also HA in an exolytic fashion. |
Unsaturated disaccharide |
Table 1. Commercially available bacterial hyaluronidases as well as chondroitinases
Treatment with bacterial eliminases converts the original uronic acid structure in the polysaccharides into an artificial structure, the 4,5-unsaturated uronic acid, 4-deoxy-α-L-threo-hex-4-enepyranosyluronic acid.

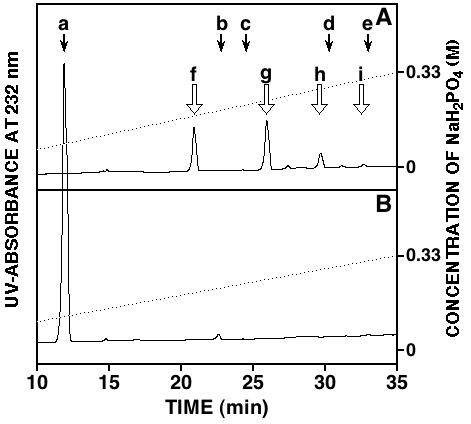
Fig. 1. HPLC profiles of the hyaluronidase digests.
The HA preparation (2 μg) was exhaustively digested with the hyaluronidase from Streptomyces hyalurolyticus (A) or hyaluronidase SD from Streptococcus dysgalactiae (B). Digests were analyzed by anion-exchange HPLC on an amine-bound silica column using a linear gradient of NaH2PO4 from 16 mM to 530 mM over 60 min. Eluates were monitored by UV-absorbance at 232 nm. Arrows indicate the elution positions of authentic unsaturated disaccharides. a, ΔHexA-GlcNAc and ΔHexA-GalNAc; b, ΔHexA-GalNAc(6-sulfate); c, ΔHexA-GalNAc(4-sulfate); d, ΔHexA(2-sulfate)-GalNAc(6-sulfate); e, ΔHexA-GalNAc(4,6-disulfate). ΔHexA, GalNAc, and GlcNAc represent 4,5-unsaturated uronic acid, N-acetylgalactosamine, and N-acetylglucosamine, respectively. Presumable positions of unsaturated oligosaccharides are shown by open arrows. f, unsaturated tetrasaccharide: g, unsaturated hexasaccharide: h, unsaturated octasaccharide: i, unsaturated decasaccharide.

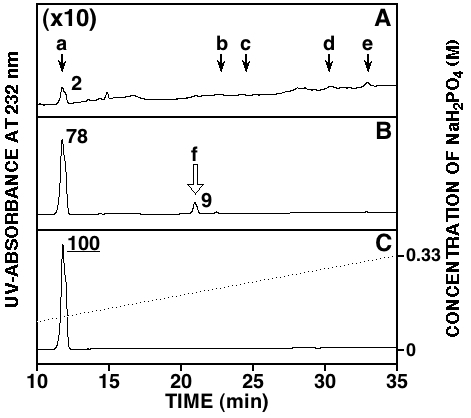
Fig. 2. HPLC profiles of the chondroitinase digests.
The HA preparation (2 μg, approximately 5 nmol as disaccharide) was digested with 5 mIU of chondroitinase ABC (A), AC-I (B), or AC-II (C) at 37˚C for 30 min. Reactions were terminated by heat treatment at 100˚C for 1 min. Digests were analyzed by anion-exchange HPLC on an amine-bound silica column, being monitored by measuring UV-absorbance at 232 nm. Arrows in panel A indicate the elution positions of authentic unsaturated disaccharides (see the legend to Fig. 1). The open arrow f in panel B shows the presumable position of the unsaturated tetrasaccharide. The chromatogram in panel A was enlarged ten times to compare it with those in panels B and C. The ratio of the major unsaturated oligosaccharides generated by enzymatic digestion was calculated based on the peak area, and is indicated in the figure taking the value obtained in the digest with chondroitinase AC-II as 100.
|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2013-12-27 12:03:18 |
- Yamagata, T., Saito, H., Habuchi, O., and Suzuki, S. (1968) Purification and properties of bacterial chondroitinases and chondrosulfatases. J. Biol. Chem. 243, 1523-1535 [PMID : 5647268]
- Ohya, T., and Kaneko, Y. (1970) Novel hyaluronidase from Streptomyces. Biochim. Biophys. Acta 198, 607-609 [PMID : 5436162]
- Hiyama, K., and Okada, S. (1975) Crystallization and some properties of chondroitinase from Arthrobacter aurescens. J. Biol. Chem. 250, 1824-1828 [PMID : 234466]
- Jandik, K.A., Gu, K., Linhardt, R.J. (1994) Action pattern of polysaccharide lyases on glycosaminoglycans. Glycobiology 4, 289-296 [PMID : 7949654]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Yamada, Shuhei,
(2013). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.26,4,2024 .
How to Cite this Work in Website:
Yamada, Shuhei,
(2013).
Bacterial glycosaminoglycan lyases to depolymerize hyaluronan.
Retrieved 26,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t156.
html source
Yamada, Shuhei,
(2013).
<b>Bacterial glycosaminoglycan lyases to depolymerize hyaluronan</b>.
Retrieved 4 26,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t156" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t156</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|