To produce an N-glycosylated protein in vitro, the extract from human hybridoma cells is programmed with mRNA encoding a protein of interest 1).
The hybridome extract is prepared from well-growing hybridoma cells, and then supplemented with ATP, GTP, creatine phosphate, creatine kinase, amino acids, calf liver tRNA and spermidine. Furthermore, to relieve phosphorylation of eIF2α due to the presence of ATP and creatine phosphate, the extract is supplemented with recombinant GADD34 and K3L. When the hybridoma extract supplemented with these reagents is incubated with synthetic mRNA encoding an open reading frame with a signal sequence, an N-glycosylated protein is synthesized co-translationally (Fig. 1). After 1-2 h of incubation, the microsomal fraction is obtained by centrifugation and is treated with a detergent to solubilize the synthesized N-glycosylated protein. |
| Category | Isolation & structural analysis of glycans |
| Protocol Name | Synthesis of glycoproteins in vitro |
Authors
 |
Imataka, Hiroaki
*
Department of Materials Science and Chemistry, and Molecular Nanotechnology Research Center, Graduate School of Engineering, University of Hyogo
Machida, Kodai
Department of Materials Science and Chemistry, and Molecular Nanotechnology Research Center, Graduate School of Engineering, University of Hyogo
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
HF10B4 (RIKEN BioResource Center, Tsukuba, Japan) |
| ● |
E-RDF medium (Kyokuto Pharmaceutical Industrial Co., Ltd., Tokyo, Japan) |
| ● |
|
| ● |
GlutaMAX (Invitrogen/Life Technologies, Carlsbad, CA) |
| ● |
Washing buffer: 35 mM HEPES-KOH, pH 7.5, 140 mM NaCl, and 11 mM glucose |
| ● |
Extraction buffer: 20 mM HEPES-KOH, pH 7.5, 45 mM potassium acetate, 45 mM KCl, 1.8 mM magnesium acetate, and 1 mM DTT |
| ● |
High potassium buffer: 20 mM HEPES-KOH, pH 7.5, 945 mM potassium acetate, 945 mM KCl, 1.8 mM magnesium acetate, and 1 mM DTT |
| ● |
Plasmids: pUC 119-T7-EMCV-IRES and pUC 119-T7-HCV-IRES |
| ● |
T7, T3 or SP6 RNA polymerase |
| ● |
mRNA synthesis kits: RiboMAX large scale RNA production system (Promega Corp., Fitchburg, WI) or mMESSAGEmMACHINE (Ambion/Life Technologies, Carlsbad, CA) |
| ● |
RNA purification column: Chroma spin+TE-30 column (Takara Bio Inc., Otsu, Japan) |
| ● |
Proteinase K (Takara Bio Inc.) |
| ● |
|
| ● |
ATP, GTP, UTP, CTP and creatine phosphate (Sigma-Aldrich, St. Louis, MO) |
| ● |
Twenty amino acids: The amino acid mixture consisting of 20 amino acids is made by mixing an equal volume of MEM amino acids solution (× 50) (Invitrogen/Life Technologies), MEM non-essential amino acids solution (× 50) (Invitrogen/Life Technologies), and 20 mM L-glutamine solution (Invitrogen/Life Technologies). |
| ● |
Creatine kinase (Roche Applied Science, Penzberg, Germany) |
| ● |
Mixture-1: Mixture-1 consists of 6 μL of GADD34 (0.2 mg/mL) and 6 μL of K3L (0.4 mg/mL) 1) 2). |
| ● |
Mixture-2: Mixture-2 (650 μL) consists of 56 μL of magnesium acetate (63 mM), 354 μL of potassium acetate (157 mM for the EMCV-IRES-dependent system or 564 mM for the HCV-IRES-dependent system), 82.5 μL of DTT (100 mM), and 158 μL of HEPES-KOH (400 mM), pH 7.5. |
| ● |
Mixture-3: Mixture-3 (180 μL) consists of 11.3 μL of ATP (200 mM), 1.1 μL of GTP (200 mM), 36 μL of creatine phosphate (1 M), 18 μL of creatine kinase (6 mg/mL), 18 μL of calf liver tRNA (9 mg/mL, Novagen), 18 μL of the mixture of 20 amino acids, and 77.6 μL of H2O. |
|
Instruments
 |
| ● |
Cellmaster Model 1700 (Wakenyaku Co., Ltd., Kyoto, Japan) |
| ● |
Mini-Bomb cell disruption chamber (Kontes Glass Co., Vineland, NJ) |
|
| Methods |
|
1. |
Preparation of hybridoma cell extract |
| 1) |
Culture HF10B4 cells at 37˚C in E-RDF medium supplemented with 10% heat-inactivated fetal calf serum and GlutaMAX (2 mM) (Note 1) using a spinner flask connected with Cellmaster Model 1700 (Note 2) with the control values of temperature (37˚C), pH (7.0), oxygen density (6.7 ppm), and stirring speed (20 rpm). |
Comment 0
|

|
| 2) |
Harvest the cells when the cell density reaches 1.0–1.5 × 106 cells/mL (Note 3). |
Comment 0
|

|
| 3) |
Wash the cells three times with the washing buffer and once with the extraction buffer. |
Comment 0
|

|
| 4) |
Resuspend the cell pellet in an equal volume of the extraction buffer (approximately 3.0 × 108 cells/mL). |
Comment 0
|

|
| 5) |
Disrupt cells by nitrogen pressure (1.0 MPa, 30 min) in the Mini-Bomb cell disruption chamber. |
Comment 0
|

|
| 6) |
Mix the cell homogenates with 1/29 volume of the high potassium buffer. |
Comment 0
|

|
| 7) |
Centrifuge twice at 700 × g for 5 min at 4˚C and recover the supernatant. |
Comment 0
|

|
| 8) |
Divide the supernatant (24–28 mg protein/mL) into aliquots, and freeze them in liquid nitrogen. |
Comment 0
|

|
| 9) |
Store the frozen aliquots at −80˚C (Note 4). |
Comment 0
|
|
|
|
2. |
Synthesis and purification of mRNA |
| 1) |
Clone the cDNA of the protein of interest with the signal sequence at the N-terminus and a tag (FLAG) sequence (optional) at the C-terminus in the pUC 119-T7-EMCV-IRES or pUC 119-T7-HCV-IRES plasmid 3). |
Comment 0
|

|
| 2) |
Digest the plasmid with a restriction enzyme 3’ downstream of the coding region. |
Comment 0
|

|
| 3) |
Synthesize RNA using the digested plasmid as the template with RiboMAX large scale RNA production system or mMESSAGEmMACHINE. |
Comment 0
|

|
| 4) |
Purify the synthesized RNAs by using Chroma spin+TE-30 column. |
Comment 0
|
|
|
|
3. |
Programming the hybridoma extract with the mRNA |
| 1) |
Preincubate the HF10B4 cell extract (7.5 μL) with mixture-1 (1.2 μL) and mixture-2 (6.5 μL) at room temperature for 10 min (Note 5). |
Comment 0
|

|
| 2) |
Add mixture-3 (1.8 μL) and mRNA (1.0 μL, 36 ng/μL final concentration). |
Comment 0
|

|
| 3) |
Incubate the mixture (18 μL, total volume) at 32˚C for 1–2 h. |
Comment 0
|
|
|
|
4. |
Purification of the in vitro synthesizes glycosylated protein |
| 1) |
Dilute the incubated mixture 20 times with phosphate buffered saline (PBS) or an appropriate buffer. |
Comment 0
|

|
| 2) |
Centrifuged at 100,000 × g for 1 h at 4˚C. |
Comment 0
|

|
| 3) |
Suspend the pellet (microsomal fraction) in PBS or the buffer and incubate it with Triton X-100 (0.2%) for 5 min on ice. |
Comment 0
|

|
| 4) |
Purify the synthesized glycoprotein by anti-FLAG column chromatography (Sigma-Aldrich) if the FLAG sequence has been appended to the C-terminus of the protein. |
Comment 0
|
|
|
| Notes |
- Addition of GlutaMAX to the culture medium helps cells grow, and leads to an enhanced protein synthesis in the cell-free systems, even when L-glutamine is already included.
- If an appropriate controlling system for culturing cells is not available, culturing cells on Petri dishes (20–25 mL medium per 150 mm dish) is recommended.
- It is very important to recover cells when they are in the logarithmic growth phase.
- The extracts can be kept at −80˚C at least for six months without a loss of translation activity.
- Preincubation with K3L/GADD34 prior to addition of mixture-3 is important, because ATP/creatine phosphate present in mixture-3 enhances phosphorylation of eIF2α .
|
| Figure & Legends |
Figure & Legends

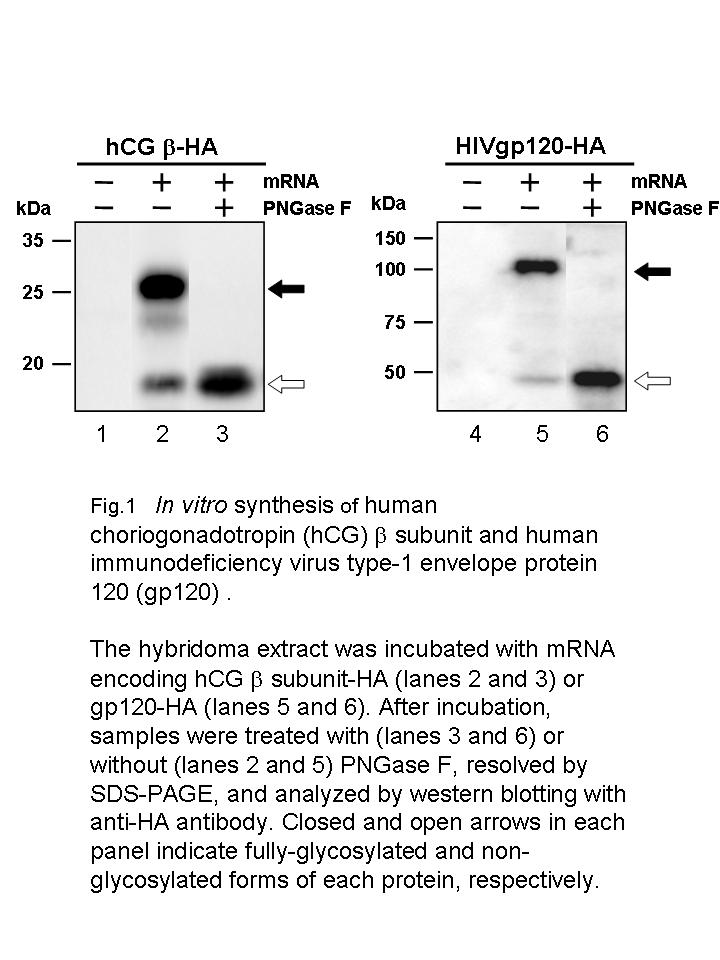
|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2014-07-30 10:38:30 |
- Mikami, S., Kobayashi, T., Yokoyama, S., and Imataka, H. (2006) A hybridoma-based in vitro translation system that efficiently synthesizes glycoproteins. J Biotechnol. 127, 65–78 [PMID : 16889861]
- Mikami, S., Kobayashi, T., Machida, K., Masutani, M., Yokoyama, S., and Imataka, H. (2010) N-terminally truncated GADD34 proteins are convenient translation enhancers in a human cell-derived in vitro protein synthesis system. Biotechnol Lett. 32, 897–902 [PMID : 20349333]
- Mikami S, Kobayashi T, Masutani M, Yokoyama S, Imataka H. (2008) A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr Purif. 62, 190–198 [PMID : 18814849]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Imataka, Hiroaki,
Machida, Kodai,
(2014). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.26,4,2024 .
How to Cite this Work in Website:
Imataka, Hiroaki,
Machida, Kodai,
(2014).
Synthesis of glycoproteins in vitro.
Retrieved 26,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t154.
html source
Imataka, Hiroaki,
Machida, Kodai,
(2014).
<b>Synthesis of glycoproteins <em>in vitro</em></b>.
Retrieved 4 26,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t154" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t154</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|