When cells are cultured in a medium that contains a radio-isotope-labeled monosaccharide, the sugar chains of the glycoproteins and glycolipids are metabolically radiolabeled efficiently. Even though there are many constraints of using radio-isotopes in the laboratory, this is a very useful method because metabolically-labeled glycans could be detected with high sensitivity by various analytical methods. Consequently, it becomes possible to deduce the structure of sugar chains by using various chromatographic methods without the use of mass spectrometer, and despite the fact that limited glycans could be prepared from the immunoprecitated glycoproteins. |
| Category | Isolation & structural analysis of glycans |
| Protocol Name | Metabolic labeling of glycans by radioactive sugars |
Authors
 |
Ohkura, Takashi
Department of Reproductive Biology, National Institute for Child Health and Development
|
| KeyWords |
|
Reagents
 |
| ● |
Glucosamine hydrochloride, D-[6-3H] (1.48-2.22TBq/mmol ; American Radiolabeled Chemicals, Inc., St. Louis, MO) |
| ● |
Culture medium specified for the cells with and without glucose (DMEM and RPMI-1640) are commercially available from Invitrogen/Life Technologies, Carlsbad, CA |
| ● |
PNGase F (Flavobacterium meningosepticum) is commercially available from Takara-Bio Inc., Otsu, Japan, Sigma-Aldrich, St. Louis, MO, (proteomics grade), and Roche Diagnostics GmbH, Mannheim, Germany |
| ● |
Sep-Pak Vac 3 cc C18 cartridge (Waters Corp., Milford, MA) |
| ● |
Dialyzed Fetal Calf Serum (FCS) |
| ● |
10 mM Tris-HCl buffered saline, pH 7.4 (TBS) |
| ● |
|
| ● |
|
| ● |
AG-50W-X8 (H+, 50–100 mesh) resin (Bio-Rad Laboratories, Hercules, CA) |
| ● |
Centrifugal ultrafilter device (e.g. Amicon Ultra-4, 10k; Merk Millipore, Billerica, MA) |
| ● |
|
| ● |
Liquid scintillation cocktail |
|
Instruments
 |
| ● |
|
| ● |
Reaction incubator (100˚C, 37˚C, 30˚C) |
| ● |
Liquid scintillation counter, such as MicroBeta TriLux 1450 (PerkinElmer, Waltham, MA) or LSC-6000 (Hitachi Aloka Medical, Ltd., Tokyo, Japan) |
| ● |
Centrifugal vacuum concentrator |
| ● |
Chromatographic systems for the further analysis |
|
| Methods |
|
1. |
Metabolic labeling of glycans by radioactive sugars |
| 1) |
Prepare ~80% confluent cells cultured in 10 cm dish under specified condition. |
Comment 0
|

|
| 2) |
Wash once with glucose-free medium containing 2% dialyzed FCS. |
Comment 0
|

|
| 3) |
Replace with 4 mL of medium containing 1.85–7.4MBq (50–200μCi) of [3H]glucosamine, 1/10 concentration of glucose and 2% dialyzed FCS. |
Comment 0
|

|
| 4) |
Incubate at 37˚C for 20–24 h under 5% CO2 atmosphere. |
Comment 0
|

|
| 5) |
Centrifuge the collected medium at 10,000 × g for 10 min, and collect the supernatant. |
Comment 0
|

|
| 6) |
Remove the radioactive components (M.W. -10kDa) with ultrafiltration device (Amicon Ultra-4), change buffer to TBS, and finally concentrate to 100–200 μL. |
Comment 0
|

|
| 7) |
Harvest the cells in PBS using a rubber cell scraper, and then centrifuge to collect cells. |
Comment 0
|

|
| 8) |
[Preparation of N-glycans]
Denature the packed cells or the solution obtained in step 6), and then digest with PNGase F according to the manufacturer's instructions at 37˚C for 18 h (See section “N-glycanase digestion”). |
Comment 0
|

|
| 9) |
Heat at 100˚C for 3 min, and then separate the released N-glycans from the reaction mixture by ultrafiltration with milli-Q water. |
Comment 0
|

|
| 10) |
Apply the filtrate as obtained above in step 9) to a Sep-Pak Vac C18 cartridge that was pre-washed with methanol and milli-Q water, and the flow-through fraction was dried using a centrifugal vacuum concentrator. |
Comment 0
|

|
| 11) |
[Preparation of O-glycans]
Transfer the residual material (0.1–0.2 mL) of the ultrafiltration device in step 9) to a 1.5 mL tube, add equal volume of 0.1 M KOH/ 2 M NaBH4 solution, and incubate at 30˚C for 15 h. |
Comment 0
|

|
| 12) |
Transfer the reaction mixture to a 15 mL tube, and add several drops of acetic acid very carefully because a large amount of H2 bubble is formed. |
Comment 0
|

|
| 13) |
When the bubbling ends, apply the reaction mixture to a Sep-Pac Vac C18 cartridge (3 cc), overlayered with 2 mL of AG-50W(H+) resin that was pre-washed with milli-Q water, and then flushed out with 10 mL of milli-Q water. |
Comment 0
|

|
| 14) |
Collect the flow-through fraction and dry it in a centrifugal vacuum concentrator. |
Comment 0
|

|
| 15) |
Add 1 mL of methanol and dry; repeat this step two more times. |
Comment 0
|

|
| 16) |
[Analysis of N-glycan and O-glycan]
Obtained N-glycans and O-glycans are subsequently available for further analysis by chromatography, such as Bio-Gel P-4 gel-permeation chromatography, Mono Q ion-exchange column chromatography, C18 reverse-phase column chromatography, Amido80 normal phase column chromatography, PGC chromatography, and serial lectin column chromatography. |
Comment 0
|
|
|
| Notes |
- Labeling with [3H]glucosamine is suitable for analyzing the side chain structure of glycans biosynthesized in the cells, because [3H]glucosamine shows the highest efficiency of incorporation into the cells and subsequent metabolical conversion to GlcNAc, GalNAc, and Gal of N-glycan and O-glycan.
- Metabolic labeling of the cultured cells with [3H]mannose for 30 min provides high mannose type glycans, lipid-linked oligosaccharide intermediate, and GPI intermediate.
- Since the sugar chains obtained from the collected culture medium are derived from both radio-labeled secreted glycoproteins and non-labeled glycoproteins from the supplemented FCS, the total glycan content might affect the behavior of the lectin column chromatography or other analysis.
|
| Figure & Legends |
Figure & Legends 
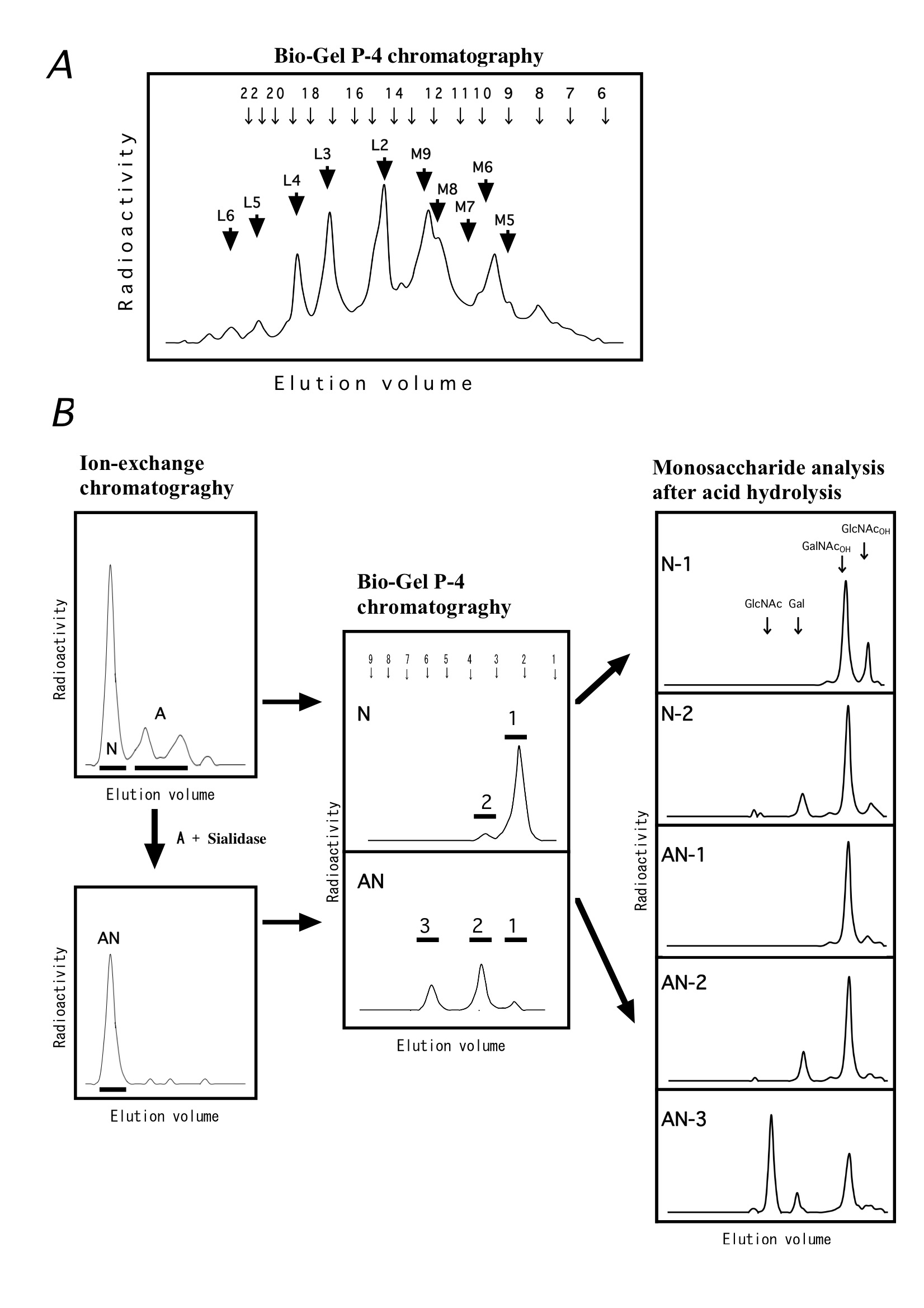
Fig. N-glycans and O-glycans derived from [3H]glucosamine-labeled Jurkat cells.
(A) N-glycans released from radiolabeled cells were reduced with NaBH4, digested with sialidase, and the obtained neutral sugar chains were then separated by using the Bio-Gel P-4 gel permeation chromatography. M5-M9 and L2-L6 correspond to Man5-9· GlcNAc· GlcNAcOT and (GalGlcNAc)2-6· Man3· GlcNAc· GlcNAcOT,, respectively. The arrows at the top of the figure indicate the elution positions of glucose oligomers (the numbers are those of glucose units). To deduce the glycan structure, use of other structural analysis method is necessary.
(B) Released O-glycans are separated to neutral sugars (N) and acidic sugars (A) by ion-exchange chromatography, and the acidic sugars (A) are neutralized by digestion with sialidase (AN). N and AN are separated using the Bio-Gel P-4 column, and the resulting peaks 1, 2 and 3 correspond to mixture of GlcNAcOH and GalNAcOH, Gal-GalNAcOH, Gal-(Gal-GlcNAc-)GalNAcOH, respectively. Right panels showed the monosaccharide analysis patterns (Shodex SP0810 column) after acid hydrolysis of the components (1-3). |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2015-05-13 16:05:47 |
- Ohkura, T., Seko, A., Hara-Kuge, S., and Yamashita, K. (2002) Occurrence of secretory glycoprotein-specific GalNAc beta 1-->4GlcNAc sequence in N-glycans in MDCK cells. J Biochem. 132, 891–901 [PMID : 12473191]
- Fukushima, K., Ohkura, T., and Yamashita, K. (1997) Synthesis of lipid-linked oligosaccharides is dependent on the cell cycle in rat 3Y1 cells. J Biochem. 121, 415–8 [PMID : 9133608]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Ohkura, Takashi,
(2015). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.25,4,2024 .
How to Cite this Work in Website:
Ohkura, Takashi,
(2015).
Metabolic labeling of glycans by radioactive sugars.
Retrieved 25,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t144.
html source
Ohkura, Takashi,
(2015).
<b>Metabolic labeling of glycans by radioactive sugars</b>.
Retrieved 4 25,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t144" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t144</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|