Glucosylceramide (GlcCer) is a common precursor for ganglio-, lacto-/neolacto-, and globo/isoglobo-series glycosphingolipids (GSLs), and galctosylceramide (GalCer), for sulfatide, GM4 (sialo-GalCer) and gala/neogala-series GSLs. The quantification of GlcCer and GalCer is somewhat difficult to perform using conventional TLC, because these simple GSLs are not easily distinguished. We describe here a new method to quantify GlcCer and GalCer by normal-phase HPLC using O-phtalaldehyde (OPA) derivatives prepared with sphingolipid ceramide N-deacylase (SCDase)1).
SCDase is an enzyme capable of hydrolyzing the N-acyl linkage of the ceramide moiety of various GSLs generating lyso-forms of GSLs and fatty acids2) 3). The enzyme hydrolyzes GlcCer and GalCer, generating glucosylsphingosine (GlcSph) and galactosylsphingosine (GalSph) which have a free NH2 group at the sphingoid moiety. The free NH2 group of GlcSph and GalSph is quantitatively coupled with OPA. OPA-GlcSph and OPA-GalSph are quantified by fluorescent detector after separation by normal-phase HPLC. This is a sensitive and reliable HPLC-based quantitative method for GlcCer and GalCer in biological samples. |
| Category | Glycolipids and related compounds |
| Protocol Name | Simultaneous quantification of glucosylceramide and galactosylceramide by HPLC |
Authors
 |
Zama, Kota
Biofunctional Lipid Biology, Frontier Research Center for Post-genome Science and Technology, Hokkaido University
Okino, Nozomu
*
Department of Bioscience and Biotechnology, Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University
Ito, Makoto
Department of Bioscience and Biotechnology, Graduate School of Bioresource and Bioenvironmental Sciences, Kyushu University
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
GlcSph (bovine) (Matreya LLC, Pleasamt Gap, PA) |
| ● |
GalSph (bovine brain) (Alexis/Enzo Life Sciences, Inc., Farmindale, NY) |
| ● |
NBD-GlcCer (Sigma-Aldrich, St. Louis, MO) |
| ● |
Psudomonas SCDase (Takara Bio Inc., Otsu, Japan) |
|
Instruments
 |
| ● |
Normal-phase column (Intersil SIL 150A-5, 4.6 × 250 mm) (GL-Sciences Inc., Tokyo, Japan) |
| ● |
Fluorescent detector (Hitachi L-7840: Hitachi, Ltd., Tokyo, Japan) |
|
| Methods |
|
1. |
Extraction of GlcCer and GalCer |
| 1) |
Add 400 μL of chloroform/methanol (1/1, v/v) and 20 μL of 1 μM C6-NBD-GlcCer (internal standard) to cell lysate (30 μL), and keep at 37°C for 2 h. |
Comment 0
|

|
| 2) |
Add 200 μL of chloroform and 150 μL of water and mix well. |
Comment 0
|

|
| 3) |
Withdraw the lower phase (chloroform layer) after centrifugation at 15,000 rpm for 5 min. |
Comment 0
|

|
| 4) |
Add 200 μL of methanol and 150 μL of water to the lower phase. |
Comment 0
|

|
| 5) |
Withdraw the lower phase after centrifugation and dry using a speed Vac concentrator. The lower phase (chloroform phase) is recovered as the GSL fraction which contains GlcCer and GalCer. |
Comment 0
|
|
|
|
2. |
SCDase treatment and OPA derivatization |
| 1) |
Add SCDase (0.6 mU in 3 μL) to the sample dissolved in 27 μL of 25 mM sodium acetate buffer, pH 5.5, containing 5 mM CaCl2 and 2.0% TritonX-100. |
Comment 0
|

|
| 2) |
Incubate at 37°C for an appropriate period. Stop the reaction with 200 μL of chloroform/methanol (1:1, v/v). |
Comment 0
|

|
| 3) |
Add water (15 μL) to the chloroform/methanol solution, mix well, and centrifuge. |
Comment 0
|

|
| 4) |
Withdraw the lower phase (chloroform phase). |
Comment 0
|

|
| 5) |
Add chloroform (200 μL) to the upper phase, centrifuge, and withdraw the lower phase. Repeat steps 3)–5) twice. |
Comment 0
|

|
| 6) |
Pool the lower phases from the 3 extractions, dry with a Speed Vac concentrator, and dissolve in 120 μL of ethanol. |
Comment 0
|

|
| 7) |
Mix the ethanol solution, and OPA reagent [0.1 mL of ethanol containing 10 mg of OPA, 20 μL of 2-mercaptoethanol, and 9.9 mL of 3 % (w/v) boric acid buffer, pH 10.5] , and incubate at 70°C for 20 min. |
Comment 0
|

|
| 8) |
Add OPA reagent (15 μL) to the ethanol solution and keep at 70°C for 60 min. |
Comment 0
|

|
| 9) |
Centrifuge the sample at 15,000 rpm for 10 min and transfer the supernatant to a glass vial. |
Comment 0
|

|
| 10) |
Inject an aliquot of sample (15 μL) into an HPLC column using an auto-sampler. |
Comment 0
|
|
|
|
3. |
|
| 1) |
Analyze the OPA-derivatized sample with C6-NBD-GlcCer (internal standard) on a normal-phase column (intersil SIL 150A-5, 4.6 × 250 mm, GL science, Japan) using n-hexane/isopropylalchol/H2O (73/26.5/0.5, v/v/v) as a mobile phase at a flow rate of 2.0 mL/min and detect the OPA-derivatives using a fluorescent detector (Hitachi L-7840) set to excitation and emission wavelengths of 340 nm and 455 nm. OPA-GlcSph and OPA-GalSph are eluted at 6.8 min and 11.3 min, respectively. |
Comment 0
|

|
| 2) |
Under the conditions described in 1), the internal standard is not detected. To quantify the internal standard, inject an aliquot of sample into the column and elute with n-hexane/isopropylalchol/H20 (44:55:1, v/v/v) at the flow rate described in 1). C6-NBD-GlcCer is eluted at 3.5 min as detected by a fluorescent detector set to excitation and emission wavelengths of 470 nm and 530 nm. |
Comment 0
|
|
|
| Notes |
- The hydrolysis of both GlcCer and GalCer by SCDase proceeds quantitatively from 5 pmol to 10 nmol under the conditions described above.
- Linearity for the determination of GlcCer and GalCer is observed from 5.0–450 pmol and 4.0–800 pmol in biological samples, respectively, which corresponds to approximately 103 to 105 RPMI cells and 5 to 80 zebrafish embryos.
- Prepare chloroform/methanol solutions just before use.
- Be careful to partition the sample with chloroform/methanol. Do not mix the lower phase with the upper phase, as this may disturb the baseline of HPLC.
- To remove contaminants such as Triton X-100 from the HPLC column, n-hexane/isopropylalchol/H20/phosphoric acid (100:60:5.7:0.3) is recommended.
- To hydrolyze GlcCer and GalCer by SCDase, extracted lipids should be completely dissolved in reaction buffer by sonication.
|
| Figure & Legends |
Figure & Legends 
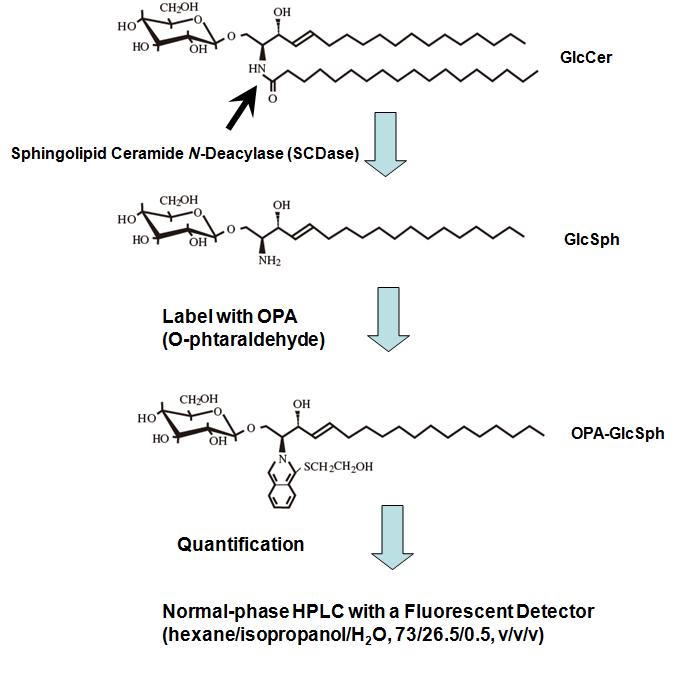
Fig. 1. Outline of the quantification of GlcCer and GalCer.
This figure was originally published in Glycobiology. 19(7):767–75. 2009 "Simultaneous quantification of glucosylceramide and galactosylceramide by normal-phase HPLC using O-phtalaldehyde derivatives prepared with sphingolipid ceramide N-deacylase" Zama K, Okino N, Ito M. et al. Oxford University Press.

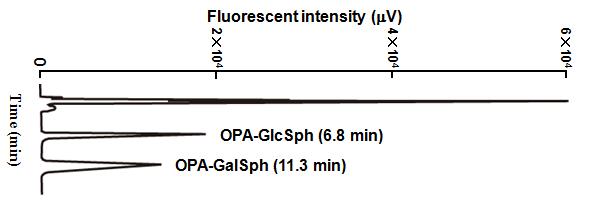
Fig. 2. Separation of OPA-GlcSph and OPA-GalSph by normal-phase HPLC.
This figure was originally published in Glycobiology. 19(7):767–75. 2009 "Simultaneous quantification of glucosylceramide and galactosylceramide by normal-phase HPLC using O-phtalaldehyde derivatives prepared with sphingolipid ceramide N-deacylase" Zama K, Okino N, Ito M. et al. Oxford University Press. |
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2016-02-04 09:50:16 |
- Zama, K., Hayashi, Y., Ito, S., Hirabayashi, Y., Inoue, T., Ohno, K., Okino, N., and Ito, M. (2009) Simultaneous quantification of glucosylceramide and galactosylceramide by normal-phase HPLC using O-phtalaldehyde derivatives prepared with sphingolipid ceramide N-deacylase. Glycobiology 19, 767–775 [PMID : 19411660]
- Ito, M., Kurita, T., and Kita, K. (1995). A novel enzyme that cleaves the N-acyl linkage of ceramides in various glycosphingolipids as well as sphingomyelin to produce their lyso forms. J. Bilo Chem. 270, 24370–24374 [PMID : 7592649]
- Furusato, M., Sueyoshi, N., Mitsutake, S., Sakaguchi, K., Kita, K., Okino, N., Ichinose, S., Omori, A., and Ito, M. (2002) Molecular cloning and characterization of sphingolipid ceramide N-deacylase from a marine bacterium, Shewanella alga G8. Journal of Biological Chemistry 227, 17300–17307 [PMID : 11827965]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Zama, Kota,
Okino, Nozomu,
Ito, Makoto,
(2016). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.19,4,2024 .
How to Cite this Work in Website:
Zama, Kota,
Okino, Nozomu,
Ito, Makoto,
(2016).
Simultaneous quantification of glucosylceramide and galactosylceramide by HPLC.
Retrieved 19,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t139.
html source
Zama, Kota,
Okino, Nozomu,
Ito, Makoto,
(2016).
<b>Simultaneous quantification of glucosylceramide and galactosylceramide by HPLC</b>.
Retrieved 4 19,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t139" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t139</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|