CL-P1 is a membrane-type collectin from placenta. The cDNA has an insert of about 2.2 kbps coding for a protein containing 742 amino acid residues. The deduced amino acid sequence shows that CL-P1 is a type II membrane protein, has a coiled-coil region, a collagen-like domain, and a CRD. It resembles type A scavenger receptors because the scavenger receptor cysteine-rich domain is replaced by a CRD. CL-P1 is expressed in vascular endothelial cells but not in macrophages. By immunoblotting and flow cytometry CL-P1 appears to be a membrane glycoprotein of about 140 kDa in human umbilical vein or arterial endothelial cells, placental membrane extracts, and CL-P1 transfected Chinese hamster ovary cells. CL-P1 can bind and phagocytose not only bacteria (Escherichia coli and Staphylococcus aureus) but also yeast (Saccharomyces cerevisiae). Furthermore, it reacts with oxidized low density lipoprotein (OxLDL). CL-P1 plays important roles in host defenses that are different from those of soluble collectins in innate immunity. |
| Category | Sugar binding proteins |
| Protocol Name | Recombinant sugar binding proteins and their functional analysis: Collectin CL-P1 |
Authors
 |
Wakamiya, Nobutaka
*
Dept. of Microbiology and Immunochemistry, Asahikawa Medical Univeristy
Ohtani, Katsuki
Dept. of Microbiology and Immunochemistry, Asahikawa Medical Univeristy
*To whom correspondence should be addressed.
|
| KeyWords |
|
Reagents
 |
| ● |
PrimeSTAR HS DNA Polymerase (Takara Bio Inc., Otsu, Japan) |
| ● |
pcDNA3.1/Myc-His A vector (Life Technologies, Inc., Carlsbad, CA) |
| ● |
Lipofectamine LTX reagent (Life Technologies, Inc.) |
| ● |
CHO-ldlA7 cells which lack functional LDL receptors |
| ● |
|
| ● |
|
| ● |
G418 (Life Technologies, Inc.) |
| ● |
Anti-Myc monoclonal antibody (Life Technologies, Inc.) |
| ● |
Anti-mouse IgG-conjugated Alexa 594 (Life Technologies, Inc.) |
| ● |
Anti-mouse IgG-conjugated Alexa 488 (Life Technologies, Inc.) |
| ● |
1,1'-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate (DiI) (Life Technologies, Inc.) |
| ● |
|
| ● |
|
| ● |
Polycationic ligands (poly(A), poly(C)) |
| ● |
Polyanionic ligands (poly(G), poly(I)) |
| ● |
|
| ● |
PBS containing 4% paraformaldehyde, pH 7.4 |
| ● |
SlowFade antifade reagent (Life Technologies, Inc.) |
| ● |
E. coli (K12 strain) BioParticles conjugated with Texas Red (Life Technologies, Inc.) |
| ● |
Staphylococcus aureus BioParticles conjugated with tetramethylrhodamine (Life Technologies, Inc.) |
| ● |
Zymosan A (Saccharomyces cerevisiae) BioParticles conjugated with Texas Red (Life Technologies, Inc.) |
|
Instruments
 |
| ● |
CO2 incubator for cell culture |
| ● |
|
| ● |
|
| ● |
|
| ● |
|
| ● |
0.2 mL in 14-mm wells of 35-mm plastic culture dishes |
| ● |
|
| ● |
10DGR desalting column (Bio-Rad Laboratories, Hercules, CA) |
| ● |
|
| ● |
|
|
| Methods |
|
1. |
Establishment of stable cell line to CL-P1 |
| 1) |
Amplify a full-length cDNA of human CL-P1 from a human placenta cDNA library by PCR using the forward primer 5′-AATGCGGCCGCACCATGAAAGACGACTTCGCAGAG-3′ and the reverse primer 5′-GCTCTAGACCGCGGTAATGCAGATGACAGTAC-3′. |
Comment 0
|

|
| 2) |
Subclone the amplified human CL-P1 cDNA into pcDNA3.1/Myc-His A vector (Life Technologies, Inc.), sequence, and transfect into CHO-ldlA7 cells using Lipofectamine LTX reagent (Life Technologies, Inc.). |
Comment 0
|

|
| 3) |
Culture the cells in Ham's F-12 medium containing 5% fetal bovine serum and 0.4 mg/mL G418 (Life Technologies, Inc.) to select CL-P1 positive clones. |
Comment 0
|

|
| 4) |
Cloning positive cells using a flow cytometer with anti-Myc monoclonal antibody (Life Technologies, Inc.) and anti-mouse IgG-conjugated Alexa 594 (Life Technologies, Inc.). |
Comment 0
|

|
|
|
|
2. |
|
| 1) |
Prepare human LDL from human plasma by stepwise sodium bromide density gradient centrifugation. |
Comment 1
|

|
| 2) |
After centrifugation, recover LDL from the fractions with densities of 1.09–1.063 g/cm3. |
Comment 0
|

|
| 3) |
Prior to oxidation, pass an aliquot of LDL through a 10DGR desalting column (Bio-Rad). |
Comment 0
|

|
| 4) |
Prepare OxLDL by the incubation of LDL (2 mg/mL) at 37°C for 24 h with 50 μM CuSO4. |
Comment 1
|

|
| 5) |
Stop the reaction by the addition of 0.25 mM EDTA. |
Comment 0
|

|
| 6) |
Label LDL and OxLDL with 1,1’-dioctadecyl-3,3,3',3'-tetramethylindocarbocyanine perchlorate (DiI) (Life Technologies, Inc.). |
Comment 0
|
|
|
|
3. |
Analysis of Lipoprotein Binding |
| 1) |
Plate CHO/CL-P1 and CHO-ldlA7 cells at densities of 3 × 104 cells/0.2 mL in 14-mm wells of 35-mm plastic culture dishes and culture in Ham's F-12 medium containing 5% fetal bovine serum with or without 0.4 mg/mL G418. |
Comment 0
|

|
| 2) |
Incubate cells at 4°C for 30 min with 5 μg/mL DiI-OxLDL in the presence or absence at 200 μg/mL of LDL and OxLDL, 10 μg/ml dextran sulfate, polycationic ligands (poly(A), poly(C)), and polyanionic ligands (poly(G), poly(I)). |
Comment 0
|

|
| 3) |
To quantify the amount of DiI-OxLDL, wash the cells and then fix with PBS containing 4% paraformaldehyde, pH 7.4, treat with 1 drop of SlowFade antifade reagent (Life Technologies, Inc.), mount, and seal. |
Comment 0
|

|
| 4) |
Observe fluorescent images with fluorescence microscope. |
Comment 0
|

|
| 5) |
Fluorescence intensity was quantified using imaging software. |
Comment 0
|
|
|
|
4. |
Analysis of Microorganism Binding |
| 1) |
Incubate CHO/CL-P1 cells at 4°C for 2 h with 1 μg/mL E. coli (K12 strain) BioParticles conjugated with Texas Red (Life Technologies, Inc.), Staphylococcus aureus BioParticles conjugated with tetramethylrhodamine (Life Technologies, Inc.), or zymosan A (Saccharomyces cerevisiae) BioParticles conjugated with Texas Red (Life Technologies, Inc.). |
Comment 0
|

|
| 2) |
After binding, fix the cells at room temperature for 20 min with 4% paraformaldehyde in PBS and stain with anti-Myc monoclonal antibody and anti-mouse IgG-conjugated Alexa 488. |
Comment 0
|

|
| 3) |
Observe fluorescent images with the system described above. |
Comment 0
|
|
|
| Figure & Legends |
Figure & Legends 
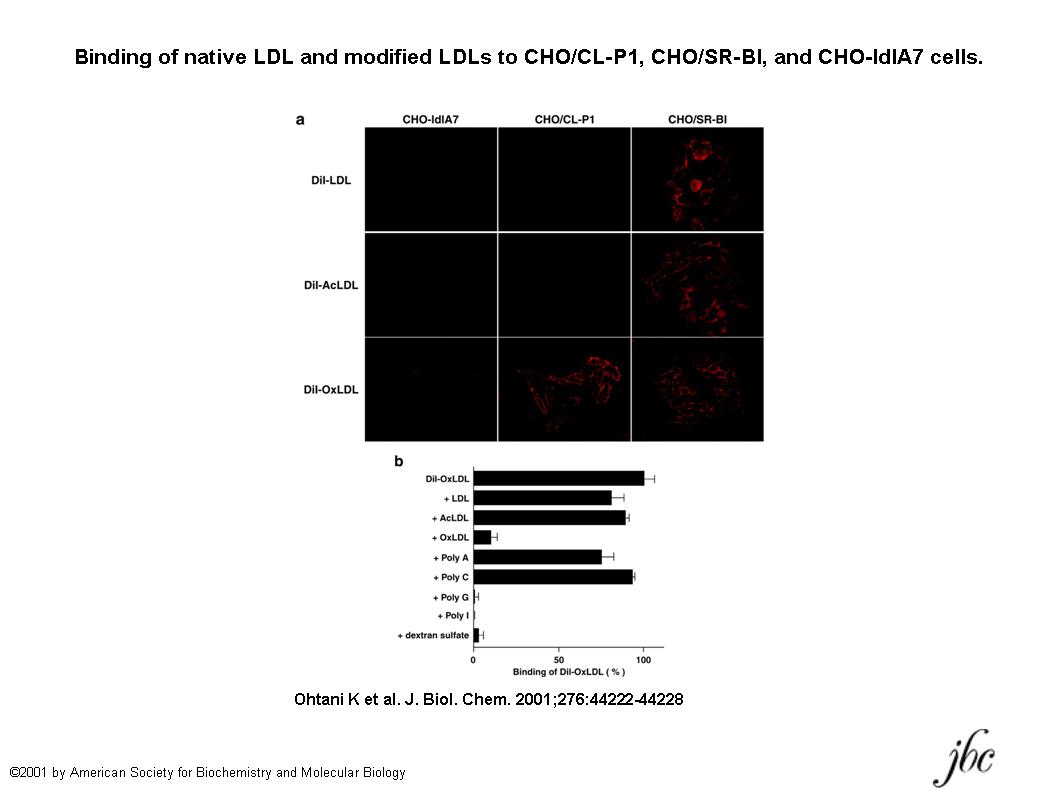
Fig. 1. Binding of native LDL and modified LDLs to CHO/CL-P1, CHO/SR-BI, and CHO-ldlA7 cells.
a, DiI-LDL, DiI-AcLDL, and DiI-OxLDL were incubated at 4°C for 30 min with CHO-ldlA7, CHO/CL-P1, and CHO/SR-BI cells. CHO/SR-BI cells were used as a positive control, and CHO-ldlA7 cells were used as a negative control. b, the binding of DiI-OxLDL was inhibited by poly(I,G), OxLDL, and dextran sulfate but not by poly(A,C), AcLDL, or native LDL. Bars indicate standard deviations.
This figure was originally published in J Biol Chem. Ohtani K. et al. "The membrane-type collectin CL-P1 is a scavenger receptor on vascular endothelial cells." 2001, 276(47): 44222–8. © the American Society for Biochemistry and Molecular Biology.

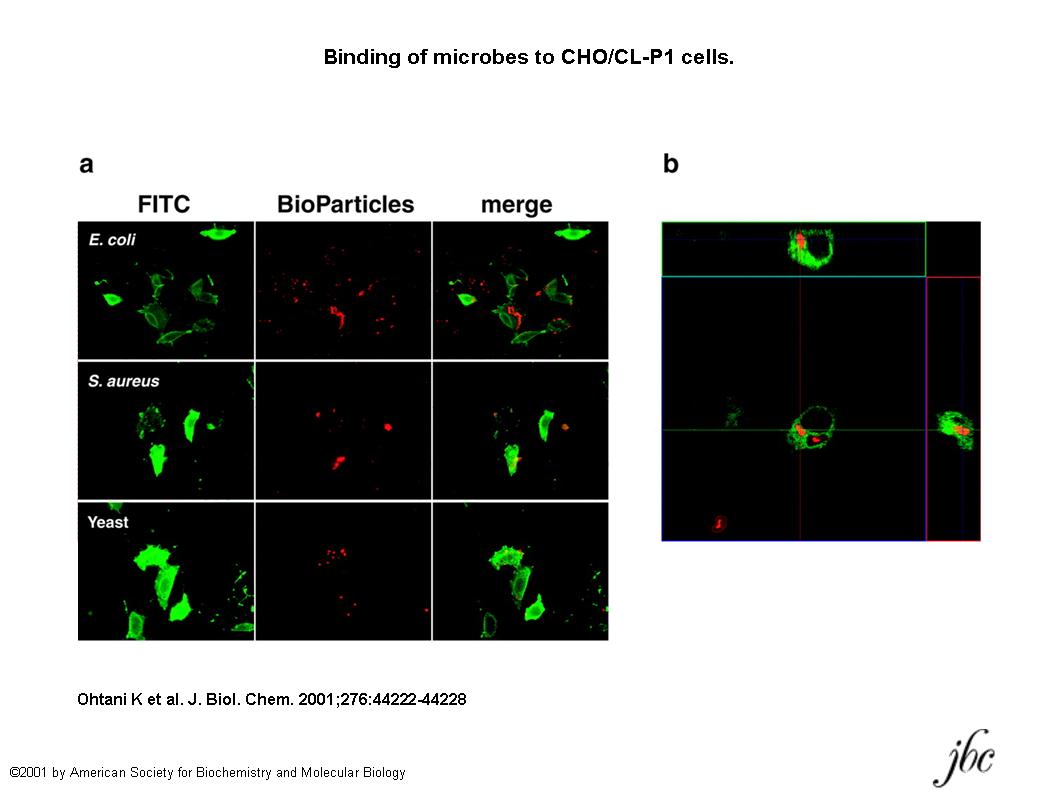
Fig. 2. Binding of microbes to CHO/CL-P1 cells.
a, photographs of CL-P1 expression and microbe binding. CHO/CL-P1 cells were stained with anti-Myc antibody and anti-mouse IgG conjugated with Alexa Fluor488. BioParticles of E. coli, S. aureus, and yeast (S. cerevisiae) conjugated with Texas Red or tetramethylrhodamine were used. b, the uptake of S. cerevisiae BioParticles by CHO/CL-P1 cells was performed at 37°C overnight under 5% CO2. After the same staining as in a, phagocytosed bioparticles were observed under a confocal laser scanning microscope.
This figure was originally published in J Biol Chem. Ohtani K. et al. "The membrane-type collectin CL-P1 is a scavenger receptor on vascular endothelial cells." 2001, 276(47): 44222–8. © the American Society for Biochemistry and Molecular Biology.
|
| Copyrights |
 Attribution-Non-Commercial Share Alike Attribution-Non-Commercial Share Alike
This work is released underCreative Commons licenses
|
| Date of registration:2014-08-08 15:02:39 |
- Ohtani, K., Suzuki, Y., Eda, S., Kawai, T., Kase, T., Keshi, H., Sakai, Y., Fukuoh, A., Sakamoto, T., Itabe, H., Suzutani, T., Ogasawara, M., Yoshida, I., and Wakamiya, N. (2001) The membrane-type collectin CL-P1 is a scavenger receptor on vascular endothelial cells. J Biol Chem. 276, 44222–8 [PMID : 11564734]
- Jang, S., Ohtani, K., Fukuoh, A., Yoshizaki, T., Fukuda, M., Motomura, W., Mori, K., Fukuzawa, J., Kitamoto, N., Yoshida, I., Suzuki, Y., and Wakamiya, N. (2009) Scavenger receptor collectin placenta 1 (CL-P1) predominantly mediates zymosan phagocytosis by human vascular endothelial cells. J Biol Chem. 284, 3956–65 [PMID : 19073604]
- Itabe, H., Yamamoto, H., Suzuki, M., Kawai, Y., Nakagawa, Y., Suzuki, A., Imanaka, T., and Takano T. (1996) Oxidized phosphatidylcholines that modify proteins. Analysis by monoclonal antibody against oxidized low density lipoprotein. J Biol Chem. 271, 33208–17 [PMID : 8969177]
|
This work is licensed under Creative Commons Attribution-Non-Commercial Share Alike. Please include the following citation
How to Cite this Work in an article:
Wakamiya, Nobutaka,
Ohtani, Katsuki,
(2014). GlycoPOD https://jcggdb.jp/GlycoPOD.
Web.23,4,2024 .
How to Cite this Work in Website:
Wakamiya, Nobutaka,
Ohtani, Katsuki,
(2014).
Recombinant sugar binding proteins and their functional analysis: Collectin CL-P1.
Retrieved 23,4,2024 ,
from https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t103.
html source
Wakamiya, Nobutaka,
Ohtani, Katsuki,
(2014).
<b>Recombinant sugar binding proteins and their functional analysis: Collectin CL-P1</b>.
Retrieved 4 23,2024 ,
from <a href="https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t103" target="_blank">https://jcggdb.jp/GlycoPOD/protocolShow.action?nodeId=t103</a>.
Including references that appeared in the References tab in your work is
much appreciated.
For those who wish to reuse the figures/tables, please contact JCGGDB
management office (jcggdb-ml@aist.go.jp).
|
|